Develop Dual-fluorescent Reporter Cell Lines with the WOLF G2
Download PDF
Introduction
Fluorescent proteins are often used in combination with fluorescence-activated cell sorting to identify and isolate successfully transfected cells or engineered reporter cell lines. A popular application of this is cell line development for disease modeling: Cells are transfected with a disease-specific mutated gene tagged with green fluorescent protein (GFP), evaluated for GFP expression, sorted as single cells, and finally, grown out into monoclonal colonies.1
GFP is the most commonly used fluorescent protein because it was the first to be isolated2,3 and is excited by a 488 nm laser, which is standard on most flow cytometer cell sorters. However, sometimes it is desirable to use fluorescent proteins with different spectral profiles or use multiple fluorescent proteins.
For example, a dual-fluorescent reporter can be engineered with red fluorescent protein (RFP) to indicate errors in gene translation, and engineered with GFP to quantify and normalize
those translation measurements.4 To develop a cell line like this, a second laser must be used, in addition to the standard 488 nm, to sort out the successfully transfected cells (GFP-like fluorescent proteins exhibit very small stokes shifts so a 488 nm laser can generally only discern one fluorescent protein5).
The WOLF G2 Cell Sorter offers a solution to do this. It is equipped with a 488 nm laser and a choice of second laser: 405, 561, or 637 nm. Here we demonstrate developing a GFPRFP dual-reporter line with the 488/561 nm configuration, while utilizing our standard industry-leading features that include gentle sorting pressure (<2 psi), sterile disposable cartridge and fluidics, and N1 single-cell dispensing.
Method
Bulk Sorting
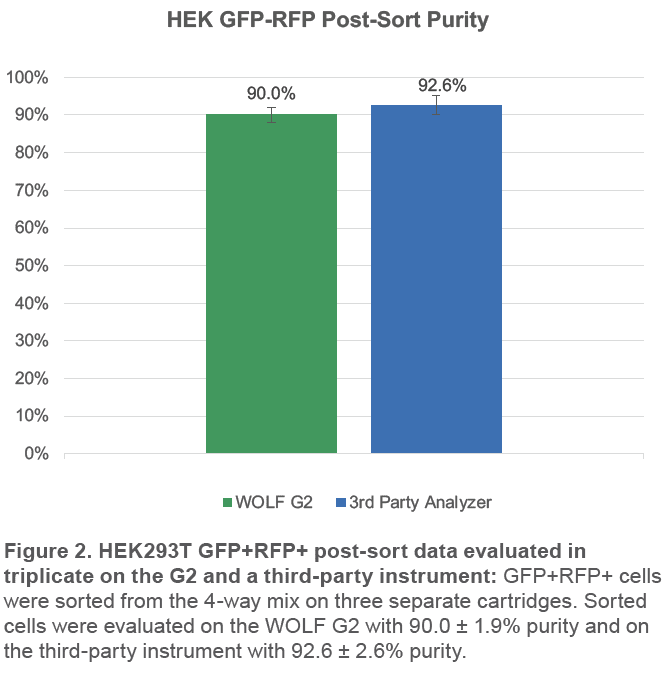
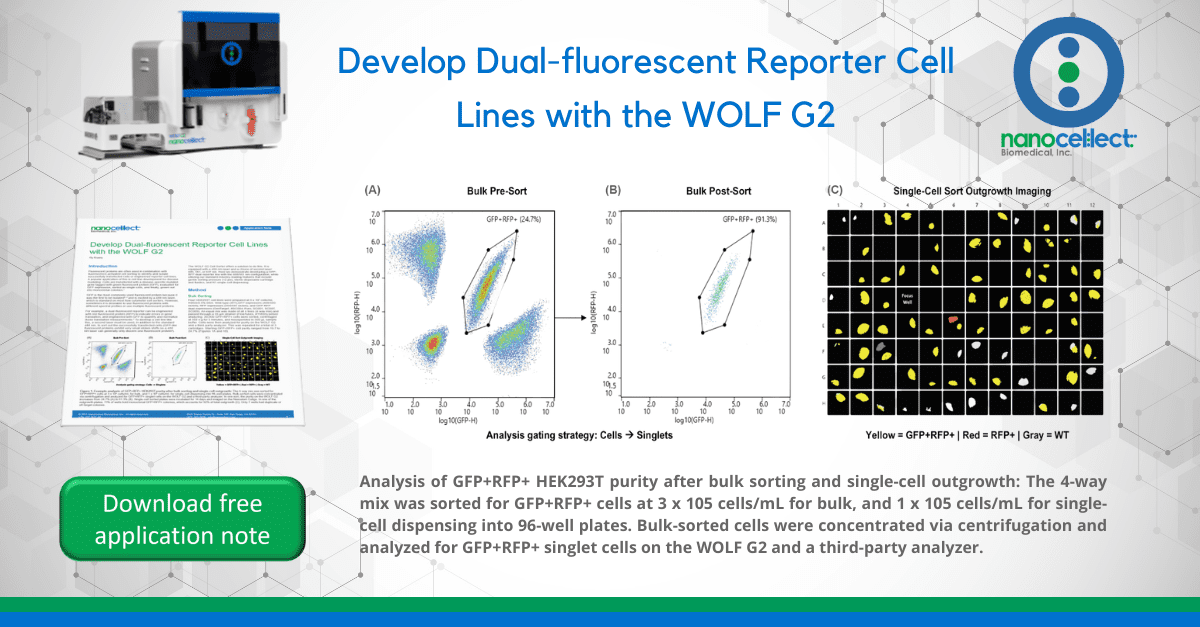
Four HEK293T cell lines were prepared at 3 x 105 cells/mL HBSS/0.5% BSA: Wild-type (WT),GFP expressors (488/509 ex/em), RFP expressors (554/591 ex/em), and GFP-RFP dual-expressors (GenTarget, #SC004-Puro, SC001, SC007, SC009). An equal mix was made of all 4 lines (4-way mix) and passed through a 35 μm strainer (FlowTubes, #T9005) before analyzing. 20,000 GFP+RFP+ cells were sorted, centrifuged at 350 x g for 5 minutes, and resuspended in 300 μL sample buffer. Cells were then analyzed for purity on the WOLF G2 and a third-party analyzer. This was repeated for a total of 3 cartridges. Starting GFP+RFP+ cell purity ranged from 19.7 to 24.7% (Figures 1A and 1B).
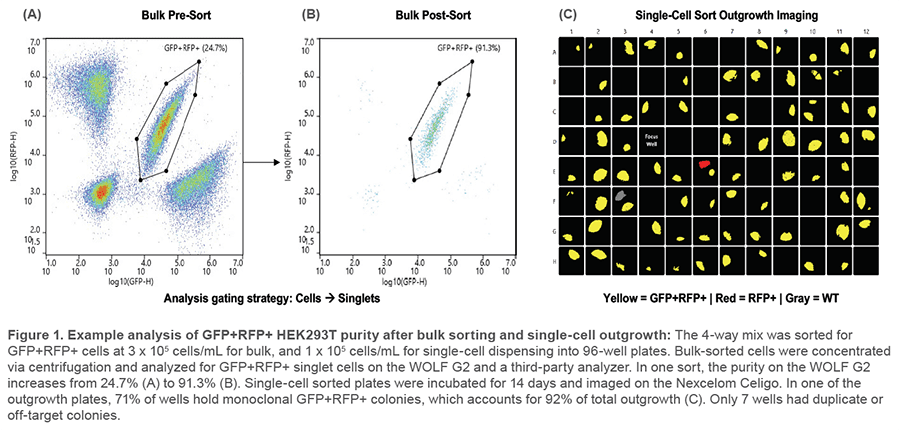
Single-Cell Sorting and Outgrowth
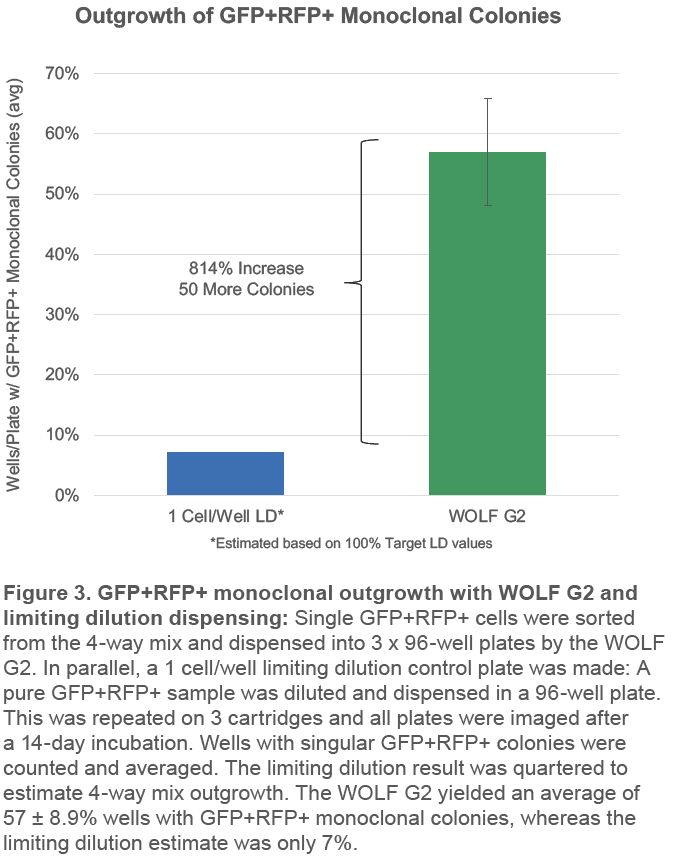
The 4-way mix was diluted to 1 x 105 cells/mL HBSS/0.5% BSA and, from it, GFP+RFP+ single cells were sorted into 3 x 96- well plates. For comparison, a 1 cell/well limiting dilution control plate was made: A pure GFP+RFP+ sample was diluted to 100 cells/mL HBSS/0.5% BSA and from that, 10 μL was dispensed into each well of a 96-well plate. All plates were pre-filled with 200 μL FluoroBrite DMEM (ThermoFisher, #A1896701)/10% FBS (GenClone, #25-514H)/4mM L-glutamine (ThermoFisher, #35050-061). For an imaging focus control, 10 μL of the 4-way mix was dispensed into the D4 well of each plate. Plates were centrifuged at 100 x g for 30 seconds and incubated at 37°C, 5% CO2 for 14 days. Plates were imaged on the Nexcelom
Celigo Image Cytometer (Figure 1C) and colony number and fluorescence were counted. Since limiting dilution counts were from a pure GFP+RFP+ sample, they were divided by 4 to estimate results from a 4-way mix (25% starting purity). This was repeated for a total of 3 cartridges.
Results
Bulk Sorting
The WOLF G2 was able to successfully purify GFP+RFP+ cells from the 4-way mix. The third-party analyzer indicates over 92% of sorted cells are GFP+RFP+; the WOLF G2 indicates a similar purity of 90% (Figure 2). The small difference in purities is likely due to variation in gating, fluidics, and optics but both still indicate high purity, > 90%.
Single-Cell Sorting and Outgrowth
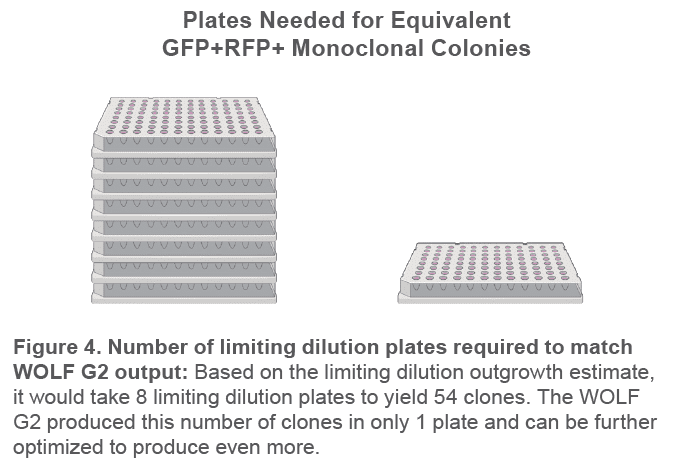
The WOLF G2 successfully yielded 57% GFP+RFP+ monoclonal colony outgrowth across 9 plates and 3 cartridges (Figure 3). This is more than 8 times as many target monoclonal colonies than the limiting dilution estimate of 7%. This means it would take 8 x 96-well limiting dilution plates to yield as many monoclonal GFP+RFP+ colonies as 1 x 96-well WOLF G2 plate (Figure 4).
Conclusion
The WOLF G2 Cell Sorter successfully sorted GFP+RFP+ ‘mock-transfected’ cells with an average purity of > 90% on multiple instruments. The higher purity on the third-party analyzer is a strong indication of success since it has very high resolution and low carryover.
The WOLF G2 also yielded an average of 57% monoclonal GFP+RFP+ colonies in 96-well plates, which was 8 times higher than the limiting dilution control estimate. The number of colonies could be increased even further by optimizing dissociation timing, culture medium, and adding a viability dye or other markers of cell health and metabolism. The 9 detection channels on the WOLF G2 make it possible to sort on more than just the dual-fluorescent reporter, all while taking advantage of the instrument’s high viability, sterility, and purity.
Acknowledgements
Thanks to Tom Chu, Bela Desai, and Adonary Muñoz for their
support on this project.
For more information, visit nanocellect.com or email [email protected]
References
1. Kim, J. J., LaGrone, A., Krasny, A., Sullivan, N., Alaynick, W., & Jose Morachis. (2021, February 19). Gentle Sorting Accelerates Generation of Genome-Edited iPSCs. Nanocellect. https://nanocellect.com/scientific-content/gentle-sorting-accelerates-generation-of-genome-edited-ipscs/
2. Shimomura, O., Johnson, F. H., & Saiga, Y. (1962). Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan,Aequorea. Journal of Cellular and Comparative Physiology, 59(3), 223–239. https://doi.org/10.1002/jcp.1030590302
3. Chalfie, M., Tu, Y., Euskirchen, G., Ward, W., & Prasher, D. (1994). Green fluorescent protein as a marker for gene expression. Science, 263(5148), 802–805. https://doi.org/10.1126/science.8303295
4. Gomes, A. C., Kordala, A. J., Strack, R., Wang, X., Geslain, R., Delaney, K., Clark, W. C., Keenan, R., & Pan, T. (2016). A dual fluorescent reporter for the investigation of methionine mistranslation in live cells. RNA, 22(3), 467–476. https://doi.org/10.1261/rna.054163.115
5. Piatkevich, K. D., Malashkevich, V. N., Morozova, K. S., Nemkovich, N. A., Almo, S. C., & Verkhusha, V. V. (2013). Extended Stokes Shift in Fluorescent Proteins: Chromophore–Protein Interactions in a Near-Infrared TagRFP675 Variant. Scientific Reports, 3(1). https://doi.org/10.1038/srep01847
APN-027