Regeneration of Plant Embryogenic Microspores Enriched with the WOLF G2® Gentle Cell Sorter
Introduction
Double haploid (DH) technology has become an integral component of commercial breeding programs in many agriculturally important crops1–4. The primary advantage of this technology is the production of homozygous haploid inducer lines in an accelerated manner1,3. The timeline differs greatly from the conventional breeding process, which requires approximately 6-10 generations of inbreeding to establish a desired cultivar1. DH methodology is used for both practical applications (e.g., breeding, mutagenesis, transformation, gene-editing) and fundamental research (e.g., biochemical, physiology, genomic studies). The process of DH production requires two steps: (1) haploid induction and (2) genome doubling, which is performed through chemical means1,2,5,6.
Microspore culture is one methodology in plant biology used to generate DH plants, which are also defined as homozygous true breeding lines1,5–10. Microspores are immature pollen grains obtained from a donor plant, such as an F1 generation1,5,6,8,9. Brassica napus, also known as canola or rapeseed, is one of the main oil crops in the world and is also a model for routine DH development via microspore culture to accelerate the breeding of new cultivars2. The microspore, rather than following the gametophytic developmental route and fertilizing the egg cell to get seed, will develop into a haploid embryo5,6,11. Several factors can influence the response of the microspore including genotype, donor plant conditions, developmental stage of the pollen grain, pretreatments, media composition, and culture conditions4–6. Although B. napus is used as a model species for microspore culture, a low frequency of microspores responds to culture. An enrichment of the culture with responding microspores would increase the efficiency of the protocol.
The WOLF G2® Cell Sorter enriches populations of interest, such as microspores, using a gentle sorting mechanism (<2psi) with low shear stress, and supports cell viability by enabling the use of culture medium as sheath. Here, we demonstrate enrichment of Brassica napus microspores and the regeneration of embryogenic microspores into haploid plants.
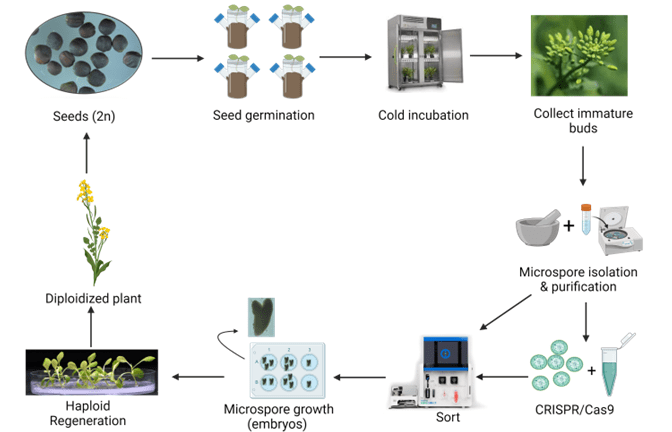
Figure 1. Microspore workflow for Brassica napus from seed to doubled haploid plant. Microspores are used for many aspects in plant biotechnology to generate DH plants that can be incorporated into breeding programs. Microspores can also be used in mutagenesis, transformation, or other genetic modification experiments with the resulting DH plants screened for the trait of interest, thereby a more rapid plant breeding methodology. This workflow was created with www.biorender.com and is based on protocols described in numerous Brassica napus publications5,6,12.
Methods
Microspore Isolation and Induction
Brassica microspores were isolated from buds based on previous methods5–7. Briefly, donor plants were grown in environmentally controlled chambers set at 20/15°C (16 h light/ 8 h dark). Just prior to bolting, the temperature was reduced to 10/5°C while maintaining the same photoperiod. The buds were collected when the microspores reached the mid-late uninucleate stage. They were sterilized in 5.25% sodium hypochlorite for 15 minutes and washed three times in sterile distilled water. The Brassica buds were then crushed in half strength B5 medium with 13% sucrose (B5-13) to release the microspores. The microspores were purified further by washing in B5-13 media three times. The number of microspores was determined using a hemocytometer and the required amount of NLN-13 (NLN medium with 13% sucrose) was added to achieve a cell density of 105 microspores per mL. Microspores were dispensed into 100 x 15 mm petri plates at a volume of 10 mL/plate and incubated in the dark at 32°C. After 72 h, the plates were transferred to 24°C.
Microscope Images
Bright field images were taken on a Zeiss Axio Observer microscope pre- and post-sort to confirm cell intactness, enrichment of desired embryogenic cells, and division and development of the microspores to embryos.
Sorting Brassica napus with WOLF G2®
Freshly isolated Brassica microspores were brought to a cell density of approximately 4.0×104 microspores per mL in NLN-13 media. The cell mixture was then slowly filtered with a sterile wide-bore serological pipet through a 70 µm nylon sieve (Corning, cat# 352340) to remove cell clumps and large particles, which could clog the cartridge13.
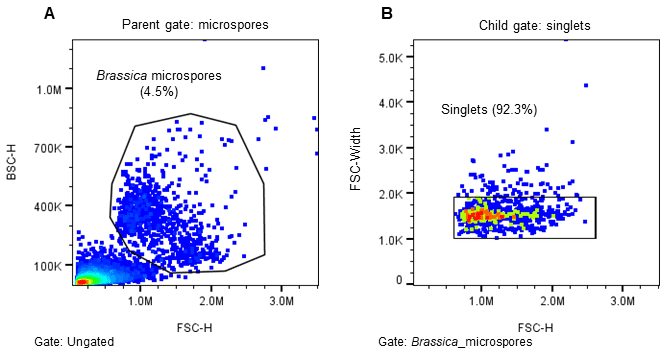
The WOLF G2 was primed and calibrated with sterile-filtered NLN-13 medium as sheath. Sterile 5.0 mL FACS tubes (Corning, cat# 352057) were placed in the tube holder intended for bulk cell sorts – three tubes corresponding to the three microfluidic sort channels named A, B, and C with B referring to the flow through and A and C for two distinct cell populations. When ready to sort, 1.5 mL of filtered microspores were transferred to a sterile 5.0 mL FACS tube. The microspores were identified on WOLF by comparing estimated cell size (FSC-H) and internal cell complexity (BSC-H) and then further separated using green autofluorescence (B525-H)(Figure 2)14–16. The gating strategy was as follows: all events expressing an FSC-H value above 1.0M and a BSC-H value above 100K are considered microspores; this is the parent gate (A). Following the parent gate is a child plot exclusive of singlet events to reduce coincidence in sorting, and it is gated to the parent (B). Third is a sub child plot displaying green positive (auto fluorescent) events solely inclusive of events from the singlets child gate (C). Lastly, (D) is a histogram representing the distribution of green auto fluorescent events in the population inclusive of only singlet events. The histogram is an alternative plot type to the scatter plot in C. Sorting for microspores herein was done using scatterplot C. Parameters and voltage gains of the sorts are listed in Table 1.

Table 1. Parameters and PMT detector gain settings for Brassica napus microspores on a WOLF G2 488/561 nm Cell Sorter. Fluorescence channel (FL) FL1 is dedicated to the 488 nm (blue) laser while FL2-FL5 channels are shared by both blue and yellow lasers, hence the “BY” nomenclature.
NOTE: To prevent cell settling and cartridge clogging, it is suggested to pause the sort and gently invert the FACS tube every 30-45 minutes. Cell settling is indicated by a drop in the number of events per second and/or a visual observation at the bottom of the sample tube.
Embryo Development and Plant Regeneration5
After sorting, the petri plates with each respective population were placed back in the incubator in the dark stationary at 24°C. Two weeks after sorting, the petri plates were checked for embryo development. Once most of the developed embryos reached the cotyledonary stage, the petri plates containing the embryos were then moved to an orbital shaker set at 70 rpm in the light at 22°C. Once the embryos turned green, they were plated on a B5 agar medium for regeneration to plants.
Results
Viable Brassica microspores were identified through successive gating using estimated cell size (FSC-H) and internal cell complexity (BSC-H). Further separation of induced and non-induced was done visualizing the green autofluorescence signal (FL1; B525); gating strategy is described in Figure 2. Microscope images post sort show isolation of viable induced and non-induced microspore populations (Figure 3).
Three populations were collected through sorting, namely High-green (HG), mid-green (MG) and low-green (LG). The low and mid-green auto-fluorescent populations represent two types of induced (embryogenic) microspores and the high-green population correlates to the non-induced microspores (Figures 2, 3).
Sorted embryogenic microspores (MG, LG) were cultured and monitored for continued cell division and embryo formation. Embryos at various developmental stages were observed two weeks after sorting (Figure 4). The timeline of development matched with the control (unsorted) Brassica microspore cultures. The heterogeneous development of induced microspores is common in the control microspore cultures as well.
Embryos turned green three days after they were transferred to an orbital shaker in the light, which was earlier than the control Brassica microspore cultures, which took the standard 7 days. Plantlets regenerated normally after the embryos were plated on B5 solid medium (Figure 5).
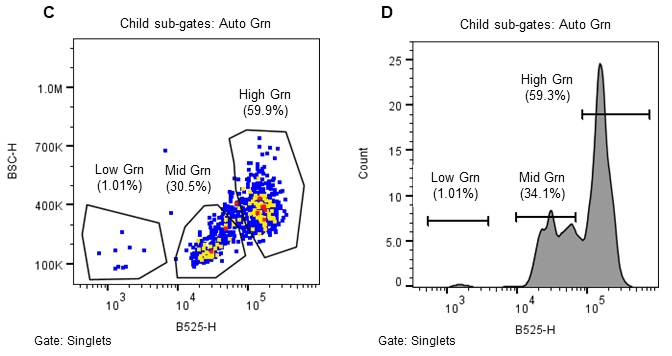
Figure 2. Sorting strategy for induced (embryogenic) Brassica napus microspores. Microspores were separated using estimated cell size (FSC-H), internal complexity/granularity (BSC-H), and green autofluorescence (FL1, B525-H) all excited by the 488nm laser. A. Parent gate (microspores), B. Child gate selecting for singlet events (gated to parent), C. Subsequent child gate selecting for green auto fluorescent positive events (gated to singlets), and D. A histogram representation of the green positive events within the child gate “singlets.”
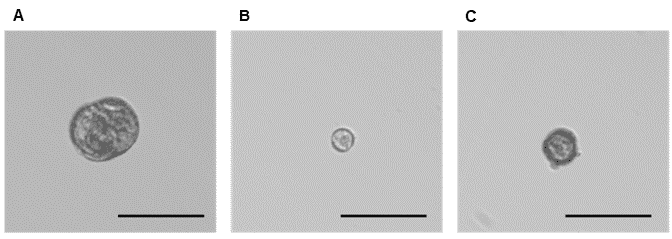
Figure 3. Phenotypes representative of three microspore populations sorted on WOLF G2. A. Low green autofluorescence, which correlates with the larger embryogenic microspore population (~1.0%), B. High green autofluorescence, which correlates with the non-embryogenic microspore population (~60.0%), and C. Mid-green autofluorescence, which correlates with an additional embryogenic microspore population (~30.0%). The scale bar on all three images is 50 µm.
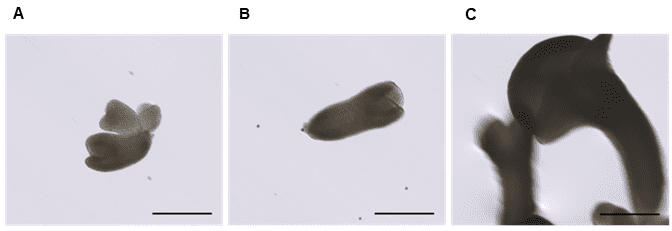
Figure 4. Embryo development two weeks after sorting on WOLF G2. A. heart-stage embryo, B. torpedo-stage embryo, and C. cotyledonary-stage embryo. Images are representative of embryogenic microspores sorted from the low green autofluorescence population in Figure 1A. The scale bar on all three images is 200 µm.
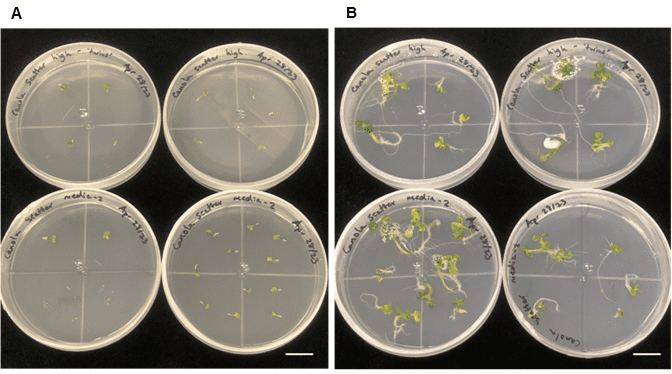
Figure 5. Freshly plated embryos and plantlet development from sorted embryogenic microspores. A. Freshly plated embryos, and B. plantlet development/regeneration two weeks after plating on solid medium. The scale bar on both images is 2.0 cm.
Conclusions
The production of homozygous true breeding lines is the goal of most commercial crop breeding systems worldwide1–4. Through traditional plant breeding processes, this can take 10+ years depending on the species1,5. Isolation of embryogenic microspores for DH production is one reliable method to create desirable cultivars in an accelerated manner1,2,4–6. Many canola (Brassica napus) breeding programs utilize this workflow to produce improved cultivars that meet canola oil standards (Figure 1). The addition of gentle cell sorting empowers scientists with a tool to select for embryogenic microspores from a heterogenous population, which further increases the efficiency of the protocol. Embryogenic microspores are desirable as they can be used in a range of workflows in both practical applications, such as DH production, and fundamental research. The WOLF G2 successfully sorted three populations of Brassica napus microspores, where the two sorted embryogenic populations successfully regenerated into plants (Figures 2-5).
Acknowledgments
Work for this application note was performed in the lab of Dr. Alison Ferrie at the National Research Council – Saskatoon, Saskatchewan, Canada*. We thank Dr. Ferrie and her team (Dr. Hai Ying Yuan and Jen Brost) for their support in the form of scientific knowledge and providing the plant material for the demonstration.
* National Research Council, 110 Gymnasium Place, Saskatoon, SK S7N 0W9, Canada website: Aquatic and Crop Resource Development Research Centre – National Research Council Canada
For more information, visit nanocellect.com or email [email protected].
References
1. Ren, J. et al. Novel technologies in doubled haploid line development. Plant Biotechnology Journal vol. 15 1361–1370 Preprint at https://doi.org/10.1111/pbi.12805 (2017).
2. Starosta, E., Szwarc, J., Niemann, J., Szewczyk, K. & Weigt, D. Brassica napus Haploid and Double Haploid Production and Its Latest Applications. Current Issues in Molecular Biology vol. 45 4431–4450 Preprint at https://doi.org/10.3390/cimb45050282 (2023).
3. Chaikam, V., Molenaar, W., Melchinger, A. E. & Boddupalli, P. M. Doubled haploid technology for line development in maize: technical advances and prospects. Theoretical and Applied Genetics vol. 132 3227–3243 Preprint at https://doi.org/10.1007/s00122-019-03433-x (2019).
4. Dunwell, J. M. Haploids in flowering plants: Origins and exploitation. Plant Biotechnol J 8, 377–424 (2010).
5. Ferrie, A. M. R. & Caswell, K. L. Isolated microspore culture techniques and recent progress for haploid and doubled haploid plant production. Plant Cell Tiss Organ Cult 301–309 (2011) doi:10.1007/s11240-010-9800.
6. Ferrie, A. M. R. & Möllers, C. Haploids and doubled haploids in Brassica spp. for genetic and genomic research. Plant Cell, Tissue and Organ Culture vol. 104 375–386 Preprint at https://doi.org/10.1007/s11240-010-9831-4 (2011).
7. Eudes, F. & Amundsen, E. Isolated microspore culture of Canadian 6x triticale cultivars. Plant Cell Tissue Organ Cult 82, 233–241 (2005).
8. Trifonova, A. & Atanassov, A. Microspore culture of winter oil rapeseed (brassica napus): II. Embryo conversion, plant regeneration and double haploid production of brassica napus l. Biotechnology and Biotechnological Equipment 11, 31–35 (1997).
9. Petrie, J. R. et al. Development of a Brassica napus (Canola) Crop Containing Fish Oil-Like Levels of DHA in the Seed Oil. Front Plant Sci 11, (2020).
10. Bhowmik, P. et al. Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci Rep 8, (2018).
11. Parra-Vega, V., Corral-Martínez, P., Rivas-Sendra, A. & Seguí-Simarro, J. M. Induction of embryogenesis in brassica napus microspores produces a callosic subintinal layer and abnormal cell walls with altered levels of callose and cellulose. Front Plant Sci 6, 1–17 (2015).
12. Corral-Martinez, P., Camacho-Fernandez, C. & Segui-Simarro, J. M. Isolated Microspore Culture in Brassica napus. vol. 2122 (Humana, 2020).
13. Dorinda JS Moellering. Protoplast isolation and enrichment using the microfluidic WOLF® Cell Sorter. Application Note 1–4 https://nanocellect.com/scientific-content/ (2022).
14. Deslauriers, C., Powell, A. D., Fuchs, K. & Pauls, K. P. Flow cytometric characterization and sorting of cultured Brassica napus microspores. Biochimica et Biophysica Acta (1991).
15. Schulze, D. & Pauls, K. P. Flow Cytometric Characterization of Embryogenic and Gametophytic Development in Brassica napus Microspore Cultures. Plant Cell Physiol vol. 39 http://pcp.oxfordjournals.org/ (1998).
16. Ochatt, S. J. Flow cytometry in plant breeding. Cytometry Part A vol. 73 581–598 Preprint at https://doi.org/10.1002/cyto.a.20562 (2008).
APN-041