Plant Nuclei Enrichment and Debris Removal with the WOLF Cell Sorter
Introduction
Plant research has led to novel discoveries in health, agriculture and the environment. Flow sorting plant cells (protoplasts) can be challenging due to their fragility and large size. In addition, some downstream applications such as single-cell DNA sequencing have cell size limitations. In instances where researchers are interested in examining the plant genome, flow sorting of plant nuclei instead of protoplasts can be a simple solution. However, obtaining and using a suspension of single plant nuclei via tissue homogenization can also provide challenges due to the large amount of irrelevant debris present in the homogenates. Cell sorters can be a valuable tool in preparation of samples of nuclei, by removing contaminating debris. However, traditional cell sorters that operate at high sample pressures have the potential to damage nuclei, particularly nuclei of species having large genomes or that are extensively endoreduplicated. The WOLF Cell Sorter uses a gentle sorting mechanism (< 2 psi) that can effectively remove debris from a target population without causing shear stress. Here, we demonstrate how the WOLF Cell Sorter can be used to remove debris and enrich for nuclei from plant tissue.
Method
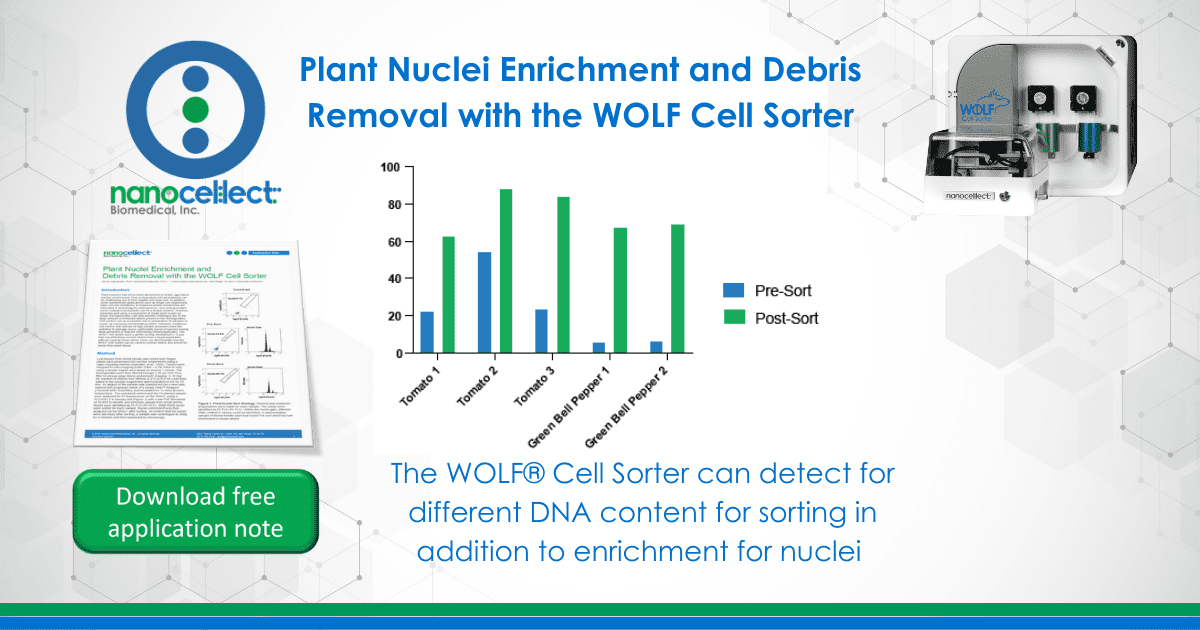
Leaf tissues from Roma tomato and Green Bell Pepper plants were processed into nuclear suspensions using a rapid chopping method (Galbraith, et al., 1983). Tissues were chopped in cold chopping buffer (PBS + 0.1% Triton-X-100) using a double-edged razor blade for around 1 minute. The homogenates were then filtered through a 20 µm Cell-Trics® filter to remove large debris and prevent clogging. A 10 mg/mL solution of DNAse-free RNAse A (2.5 µL/0.5 mL) was then added to the nuclear suspension and incubated on ice for 10 min. An aliquot of the sample was transferred into a new tube, stained with propidium iodide (PI) Ready FlowTM Reagent (ThermoFisher Scientific), and incubated for 15 mins at room temperature. The unstained control and the PI-stained sample were analyzed for PI fluorescence on the WOLF, using a FL2-H/FL3-H density plot (Figure 1) with a low FSC threshold of 10,000 to identify and eliminate signals from small debris. Nuclei were identified as PI-FL2+/PI-FL3+. 3000-5000 nuclei were sorted for each sample. Nuclei enrichment was then analyzed on the WOLF after sorting. To confirm that the nuclei were still intact after sorting, a sample was centrifuged at 300g for 5 minutes and then examined by microscopy.
Results
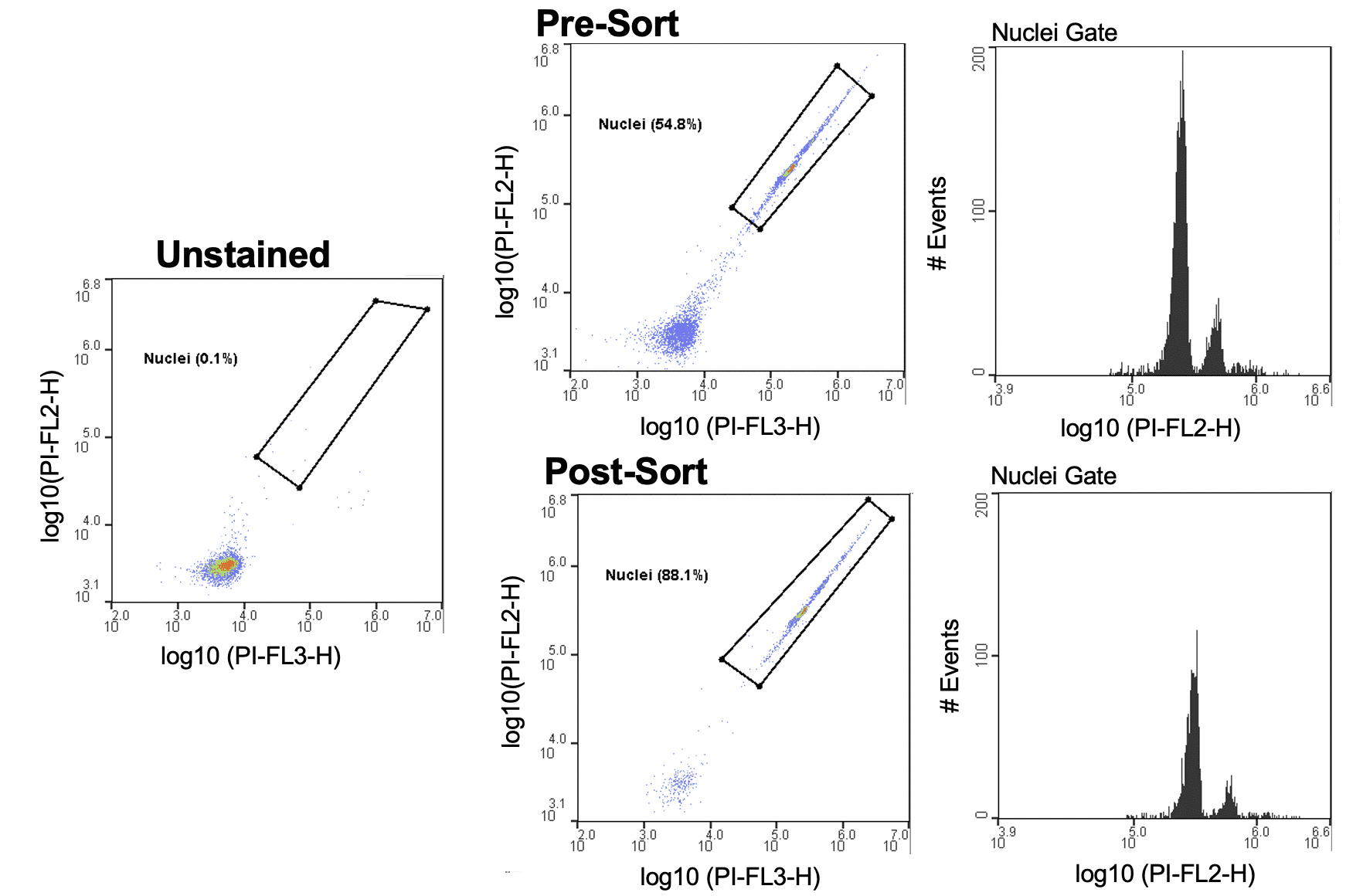
Plant nuclei were identified using an unstained sample and FL2/FL3 parameters on the WOLF. Within the PI+ nuclei gate, a histogram plot with the PI-FL2 parameter shows discrete peaks of nuclei with different PI intensities (Figure 1). These peaks represent different discrete amounts of nuclear DNA content, corresponding to nuclei at different stages of the cell division cycle and therefore having different C-values, 2C and 4C in this case (Galbraith et al 1983, Galbraith 2009). Plant debris from tomato and green bell peppers were considerably reduced post-sort, with an average 3-fold decrease (Figure 2A). Interestingly, even using an unconventional PBS-based chopping buffer, the plant leaf nuclei also appeared intact and undamaged during analysis and following post-sorting (Figure 2B).
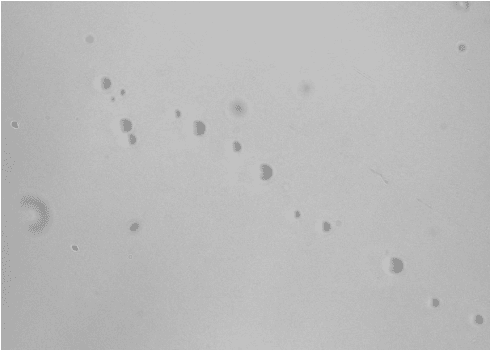
Conclusion
Enriching for plant nuclei can be challenging due to their fragility and the significant amount of debris present in the nuclei suspension. The chopping method (Galbraith, et al., 1983) is a rapid and gentle way to release nuclei from plant tissues with minimal damage; however, there is still plant debris left in the homogenate. Results from these experiments show that the WOLF Cell Sorter can serve as a robust clean-up step to remove debris and enrich for nuclei. In addition to sorting for all nuclei in the sample, the WOLF was also sensitive enough to detect different DNA content C-values within the population of nuclei. This allows sorting for nuclei with different DNA content for downstream cell cycle studies. In conclusion, without sacrificing the quality of nuclei, the WOLF Cell Sorter is an effective and gentle way to remove debris from plant nuclei samples.
References
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, and Firoozabady E (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220(4601):1049–1051. doi:10.1126/science.220.4601.1049
Galbraith DW (2009). Simultaneous flow cytometric quantification of plant nuclear DNA contents over the full range of described angiosperm 2C values. Cytometry Part A 75(8):692–698. doi:10.1002/cyto.a.20760
Galbraith DW, Janda J, and Lambert GM (2010). Multiparametric analysis, sorting, and transcriptional profiling of plant protoplasts and nuclei according to cell type. In: Hawley T., Hawley R. (eds) Flow Cytometry Protocols. Methods in Molecular Biology (Methods and Protocols) 699:407–429. Humana Press. doi:10.1007/978-1-61737-950-5_20
APN-013

