Induced Pluripotent Stem Cell Preparation for 10x Genomics Applications
Introduction
Induced Pluripotent Stem Cells (iPSCs) are adult cells, typically collected from the skin or blood, that are genetically reprogrammed to have embryonic properties. iPSCs have made a dramatic impact in biological research where they are used to study tissue and organ development, drug discovery, and a wide range of diseases. Notably, iPSCs can be derived from affected patients, such as Alzheimer’s, to produce disease-in-a-dish models. Furthermore, single-cell genomics, such as single-cell RNA sequencing (scRNA-seq), has become a valuable tool to study the diversity and heterogeneity of cells contributing to a phenotype. Transcriptional profiling of pluripotent cells is of extreme importance to scientists because it facilitates the discovery of disease development at the molecular and cellular level. iPSCs are fragile cells and mechanical stress can obscure or skew the scRNA-seq results by activating genes related to cell stress or differentiation, rather than the process of interest. Therefore, it is important to minimize mechanical stress when handling cells. Furthermore, the quality of cDNA libraries is improved by reducing dead cells and debris prior to cDNA generation and library preparation. Fluorescence activated cell sorting is a common technique used by researchers to enrich specific cell populations while avoiding dead cells and debris. However, conventional cell sorting methods that use a high-pressure droplet-based sorting cause mechanical shear stress on cells; inducing gene expression changes and/or RNA degradation.
The WOLF® Cell Sorter is a gentle, low-pressure (<2 psi) microfluidic cell sorter. Using a low-pressure cell sorter reduces the shear stress on cells to enable healthier cells and higher RNA integrity post-sort. The WOLF also uses a disposable microfluidic cartridge so that everything the sample and sheath fluid touches is sterile, disposable, and free of sample-to-sample contamination. To demonstrate how this gentle, sterile cell sorter affects single-cell RNA transcriptomics in a 10x Genomics workflow, we compared sample preparation in unsorted iPSCs to cells sorted on either the NanoCellect WOLF or BD FACSAriaTM II.
Method
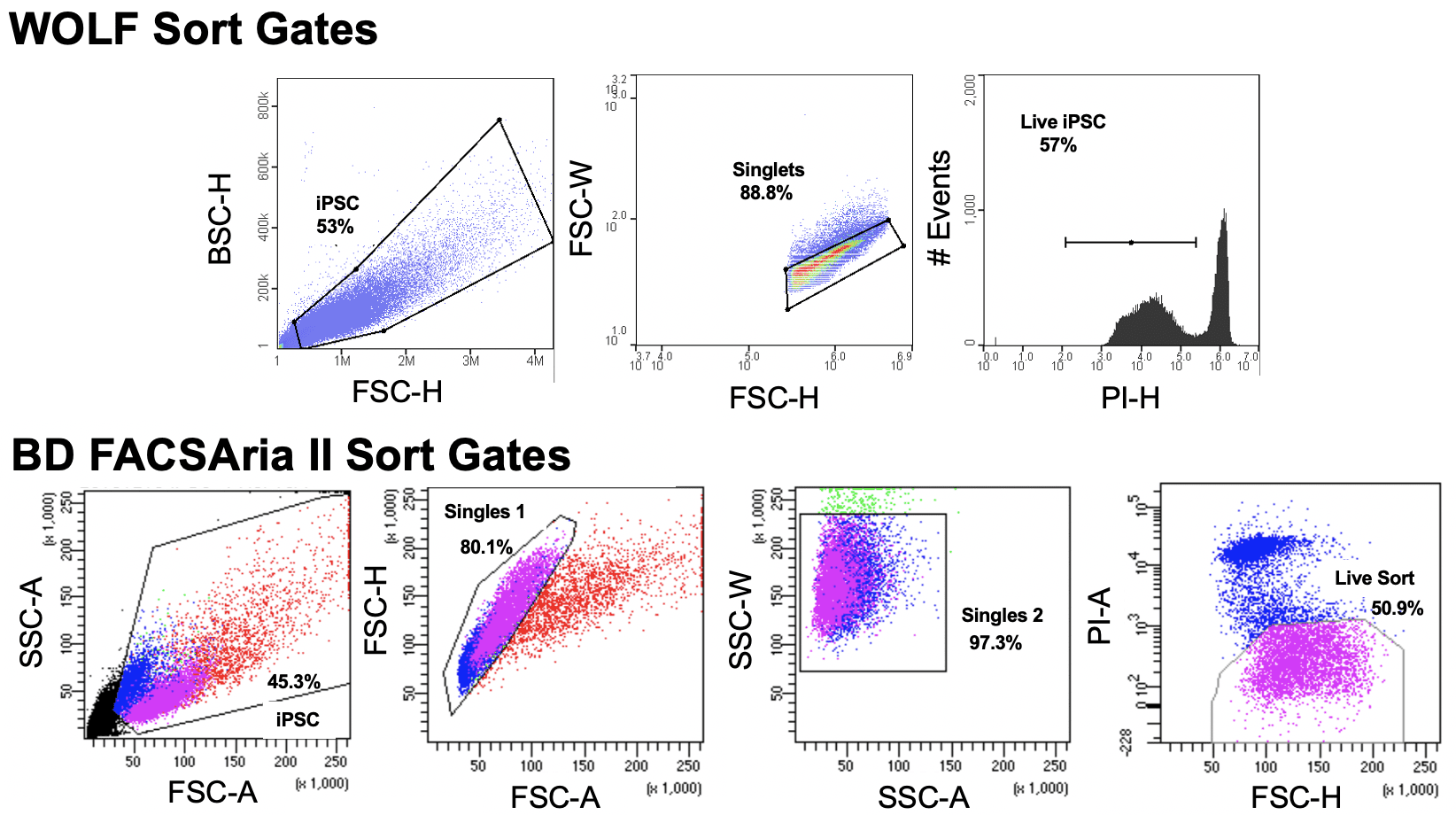
Reprogrammed iPSCs from human skin fibroblasts (Coriell Institute, GM23338) were divided into three samples: 1) unsorted control; 2) cells sorted on a conventional high-pressure droplet sorter, the BD FACSAria II; and, 3) cells sorted on the low-pressure microfluidic WOLF Cell Sorter. Prior to sorting, propidium iodide (PI, ThermoFisher #R37169), was used to identify dead cells. On the BD FACSAria II, iPSCs were sorted by the Sanford Burnham Prebys Flow Core using a 100 µm nozzle and pressure reduced to 20 psi. On the WOLF, PBS+ 0.05% BSA was used as the sheath. WOLF sort gates were set to exclude debris using BCS-H/FSC-H, and to exclude doublets with FSC-H/FSC-W. Live cells were gated as the PI-negative population. BD FACSAria II sort gates were set to exclude debris using SSC-A/FSC-A, and to exclude doublets with FSC-H/FSC-W and SSC-W/SSC-A. Live cells were also identified as the PI-negative population. Sort plots are shown in Figure 1.
50,000 cells were sorted on both the BD FACSAria II and NanoCellect WOLF into a FACS tube precoated with 0.5 mL of PBS+0.05% BSA to increase cell recovery. All samples were then centrifuged and re-suspended at ~1000 cells/µL. An estimated 10,000 cells were then loaded into the 10x Chromium Controller. cDNA libraries were generated using the Chromium Next GEM Single Cell 3’ Library Construction Kit v3.1 and sequenced with the Illumina® NovaSeq 6000 Sequencing System. All samples were analyzed with the 10x Genomics Cell Ranger pipeline to evaluate the sample and sequencing quality.
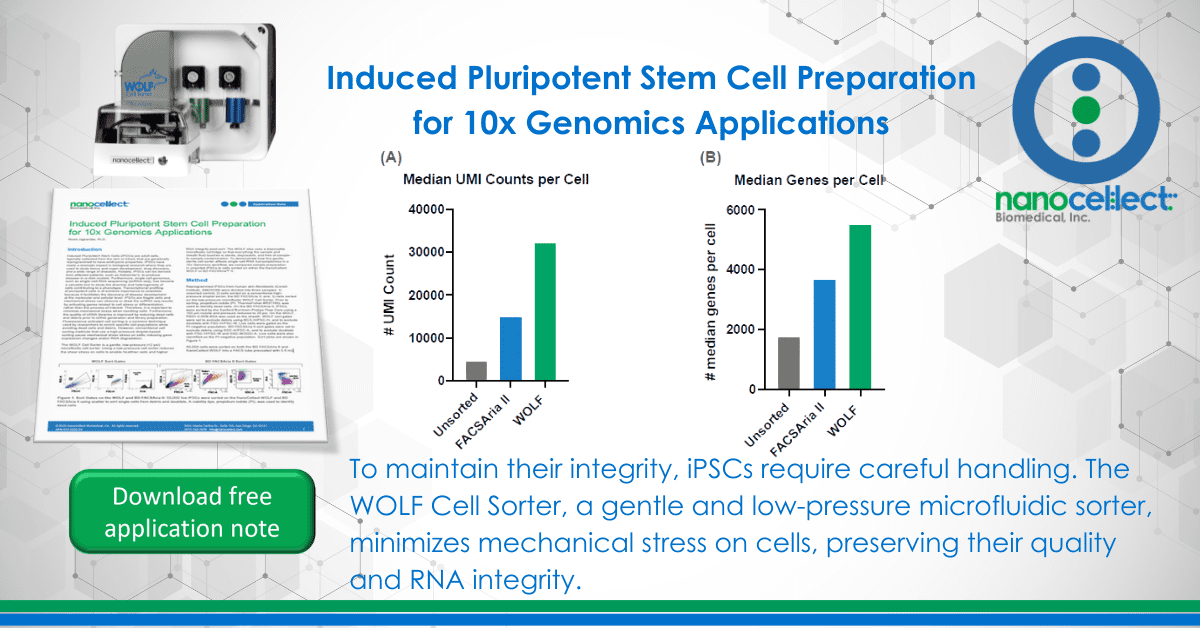
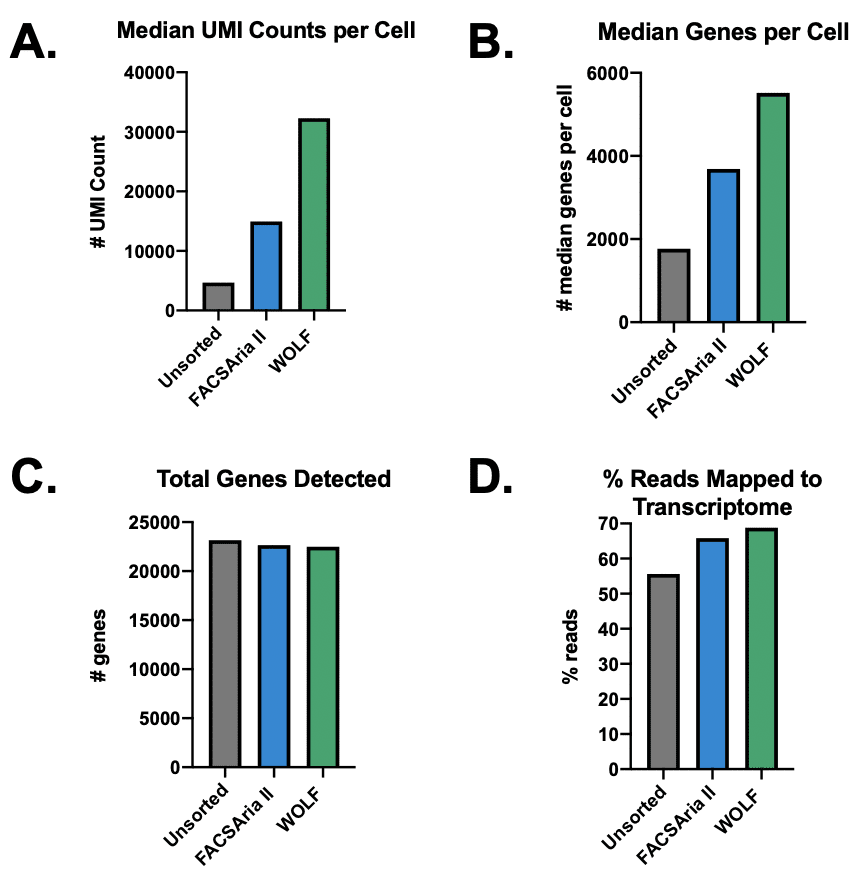
Results
The Cell Ranger summary produces several key metrics about the RNA and sequencing quality of a sample. The WOLF sorted sample ‘Median UMI Counts per Cell’ were increased nearly 7-fold increase (32,239) over the unsorted population (4,668) and 2-fold over the BD FACSAria II sorted population (14,949) (Figure 2A). A similar result was observed for ‘Median Genes per Cell’, where the WOLF had a 3-fold increase over the Unsorted cells, while the BD FACSAria II sorted sample had only 2-fold more than the Unsorted cells (Figure 2B). These results reveal that a higher library complexity is achieved when debris and dead cells are removed from the sample. Interestingly, we found a similar number of ‘Total Genes Detected’ in all three conditions (Figure 2C), perhaps due a ceiling effect of sampling so many cells. Lastly, the ‘% Reads Mapped to Transcriptome’ shows the percentage of the reads that uniquely map to a specific gene. A low percentage could indicate the presence of fragmented RNA that cannot be unequivocally mapped. Our results show an improved percentage of reads that mapped to the transcriptome in both the Aria (65.8%) and WOLF (68.8%) after sorting (Figure 2D).
Conclusion
This experiment demonstrates that using a cell sorter upstream of your 10x Genomics applications results in improved library complexity. Although library complexity was improved in the both the WOLF and Aria sorted sample, it was more improved in the WOLF sample, enabling you to get more data per cell. Furthermore, even though these results showed that there was a similar number of genes detected, there was still a higher percentage of reads that mapped uniquely to a specific gene. This suggests that there was less contamination in the sorted samples, which means you can have more confidence in the data. In summary, these results show that the WOLF serves as a robust cell sorter to use when preparing your samples for the 10x Genomics applications.
APN-010


