Epigenomic Analysis of Sorted Retinal Ganglion Cells using ATAC-seq
Background
Glaucoma is a group of diseases with diverse molecular mechanisms of pathogenesis, all of which converge on a common pathway leading to typical optic nerve damage, loss of retinal ganglion cells and, consequently, loss of vision. Retinal Ganglion Cells (RGCs) are the brain’s only visual connection to the outside world and despite numerous studies, there is not yet successful therapy to prevent their loss in glaucoma. Several studies have identified nonsynonymous coding variant SIX6-rs33912345 His141Asn as strongly associated with glaucoma. To directly assess the functional relevance of Six6-141His versus Six6-141Asn variants in eye function, transcriptomic and epigenetic changes in RGC isolated from mouse strains expressing the “protective” and “risk” SIX6 allele were assessed. A murine model of glaucoma was established by raising Intraocular Pressure (IOP) which serves as the mechanism of RGC injury.
Methods
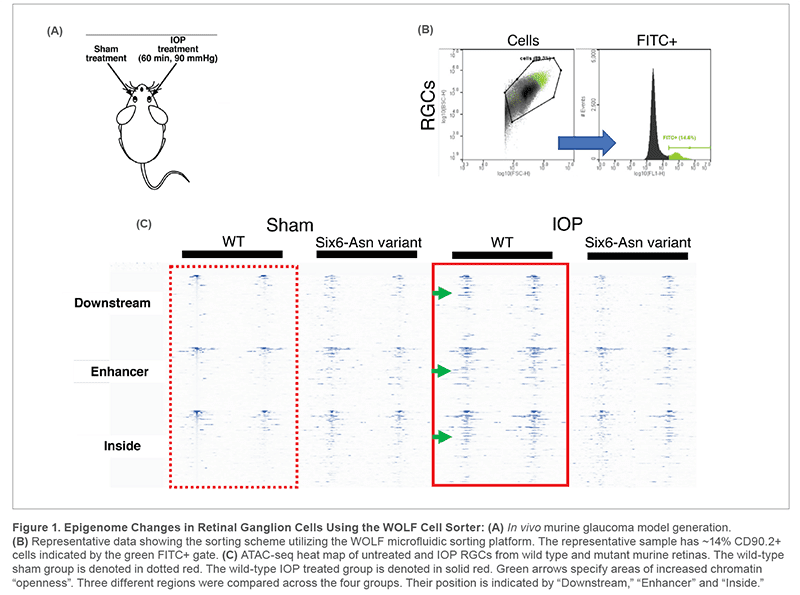
Unilateral elevation of IOP was achieved by instilling the aqueous chamber of the eye with Balanced Salt Solution through an IV infusion set. The level of IOP was maintained for 60 minutes, controlled by IOP measurements using a veterinary rebound tonometer (Tonovet). Retinas were isolated and dissociated using papain solution according to the laboratory protocol. The resulting cell suspension was stained with FITC-conjugated antibody against CD90.2 protein expressed specifically on RGCs. CD90.2 positive cells were then sorted on the NanoCellect WOLF Cell Sorter to gently sort the very fragile RGCs. Finally, ATAC-seq protocol was applied on cell suspensions isolated from both the wild-type and variant strain, which were either sham and IOP treated.
Results
Traditionally, RGCs have been difficult to sort due to their fragile nature. Now, the NanoCellect WOLF Cell Sorter offers an opportunity to sort sensitive cell types and prepare a higher quality sample than conventional practices. The NanoCellect WOLF Cell Sorter identified and separated on average 11% of CD90.2 positive cells in each genotype, before and after the IOP elevation. After completing ATAC-seq protocol, differential chromatin accessibility patterns were observed between the wild-type sham and IOP treated groups. These alterations to chromatin accessibility can be used to assess nucleosome mapping, transcription factor occupancy, as well as identify ectopically active regions in pathological conditions. The NanoCellect WOLF Cell Sorter facilitates the preparation of population-specific cell suspensions such as neurons suitable for advanced genomic analyses, like ATAC-seq.
APN-002
