Combining Microbubbles and Cell Sorting for Quick, Gentle Isolation of Unique Cell Populations
Introduction
Sample preparation is a critical step in any scientific workflow. To ensure high quality results from downstream applications such as single-cell sequencing, it is imperative that the sample be pure and unaltered by harsh retrieval techniques. Rare or limited cell populations can be especially challenging to study and often require an additional enrichment step to ensure that enough cells of interest are analyzed to provide meaningful data. While fluorescence activated cell sorting (FACSTM) has been the gold standard of cell separation methods, it can be time consuming when isolating cell populations with low frequency. Furthermore, a long sorting step can lead to a significant amount of cell death by the time enough cells are collected for downstream applications. There is a strong need for efficient, rapid, and gentle sample preparation methods to keep up with the advancements in technology for assessing numerous aspects of cellular and molecular biology.
Regulatory T cells (Tregs) represent a rare and important cell population in the immune system. They constitute 4-10% of the total CD4+ T cell population and only 1-5% of all peripheral blood mononuclear cells (PBMCs). Tregs play an important role in the immune system by regulating other cells in the immune system. Specifically, Tregs play essential roles in preventing excessive immune responses and autoimmune diseases. Clinically, Tregs have been targeted to prevent graft rejection, autoimmune diseases, and most recently, cancer. Since Tregs represent a critical component of the immune system and its response to a variety of disease states, it is vital to understand Tregs function at a molecular level. A crucial step in investigating Tregs is isolating enough cells for downstream analysis using separation methods that do not damage the cells or alter their biology. Because current isolation techniques can be time consuming and induce stress on cells, development of a gentle and efficient method for obtaining a high purity of a small target cell population for downstream analysis is critical. This application note demonstrates how to successfully isolate cells of low abundance, while saving time and maintaining high viability, through a combination of Akadeum’s microbubble based CD4+ T cell enrichment and subsequent Treg isolation using NanoCellect’s WOLF® Cell Sorter.
Microbubble Technology
Akadeum’s Buoyancy-Activated Cell Sorting (BACS™) microbubble technology represents a breakthrough in cell separation. The core of each microbubble is a hollow glass microsphere which allows researchers to leverage the power of gravity for quick and gentle removal of captured cells from a complex mixture. Because gravity is the driving force behind the microbubbles, they provide consistent results without requiring extraneous equipment like magnets or columns, which enables reliable separations regardless of sample volume or container shape or size.
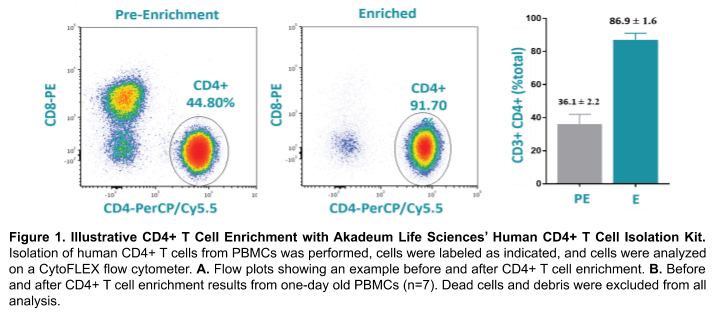
Akadeum’s Human CD4+ T Cell Isolation Kit utilizes streptavidin-coated microbubbles and a biotinylated antibody cocktail to selectively label, capture, and remove unwanted cell populations. Unwanted cells are first labeled with the provided antibodies and then bound to the streptavidin-coated microbubbles using a fast and gentle mixing step. An optional centrifugation step pellets the unlabeled CD4+ T cells, while the unwanted cells are depleted by carrying them to the top of the vessel via attachment to the microbubbles. Vacuum aspiration removes microbubbles, microbubble-bound cells, and supernatant, leaving a highly enriched population of CD4+ T cells (Figure 1). This rapid, gentle and efficient protocol considerably reduces sort times thereby lowering costs and improving cell viability.
Microfluidic Cell Sorting
The WOLF Cell sorter is a novel microfluidic cell sorter that gently sorts cells at less than 2 psi without inducing shear stress. The WOLF is powered by a 488nm laser that can detect fluorescent proteins and dyes in the green (500-550nm), yellow (565-605nm) and red (665+nm) wavelengths. Unlike traditional cell sorters, the WOLF also reduces biohazards with a disposable microfluidic cartridge so everything that the sample and sheath fluid touches is sterile, disposable, and free of sample-to-sample contamination. Although traditional cell sorters are capable of higher sorting speeds, they also exert high amounts of pressure (10-70 psi) on cells. This type of shear stress can leave cells in a stressed state that can cause high amounts of cell death and gene expression changes. The WOLF, however, can accurately and gently sort cells at 300 cells/second so that the results from experiments more accurately reflect experimental conditions with minimal sorting artefacts.
Methods
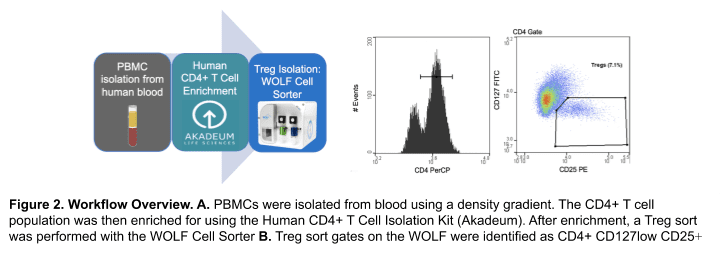
One-day old blood from each donor was used (San Diego Blood Blank). PBMCs were then isolated using a density gradient prior to enrichment. CD4+ T cell enrichment from each donor was done using the Human CD4+ T cell Isolation Kit (Akadeum #13210-130). The enriched CD4+ T cell sample was then stained with a Treg panel: CD4-PerCP (BioLegend #317431), CD25-PE (BioLegend #302605) and CD127-FITC (BioLegend, #351311). Cells were then resuspended at a concentration of 2-3 x 105 cells/mL before analysis and sorting on the WOLF. DPBS + 0.02% EDTA + 2% FBS was used as the sorting buffer and sheath on the WOLF. Sort gates on the WOLF were identified as Tregs by the CD4+ CD127low and CD25+ gate (Figure 2B). Dead cells and debris were excluded by the BSC/FSC scatter plot and doublets were removed through the FSC-Width/ FSC-Height scatter plot. 1 x 104 Treg cells were then sorted on the WOLF and the purity was accessed by flow cytometry (Acea NovoCyte). Viability was measured by the SSC/FSC plot and validated with SYTOXTM Green Ready FlowTM Reagent (ThermoFisher, #R37168) at pre-enrichment, post-enrichment, and post-sort for Donor 4.
Results
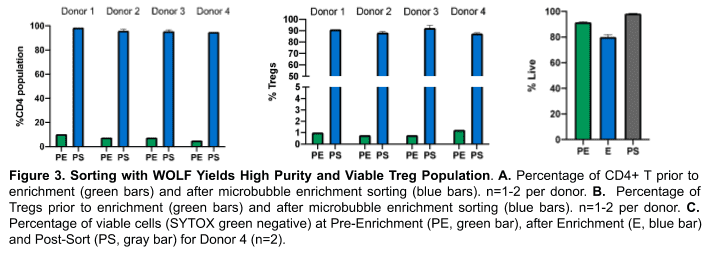
Microbubbles and microfluidic flow cytometry were used to enrich Treg cells. After enrichment, the CD4+ T cell population made up 95-98% (Figure 3A) of cells within the lymphocyte gate and Tregs within the lymphocyte population went from an average of 1% to 85-94% (Figure 3B).
Dead cells were eliminated by identifying live cells with a combination of scatter and a fluorescent viability dye (Figure 3). Pre-enrichment, an average of 91% of the cell population was viable, after enriching for the CD4+ T cell population, there was a slight reduction (80%) in the percentage of live cells (Figure 3C). However, after sorting out dead cells and debris with the WOLF, nearly all (98%) of the sorted cells were viable (Figure 3C).
Conclusion
Using Akadeum’s quick and easy CD4+ T cell enrichment microbubbles before proceeding with Treg isolation using NanoCellect’s WOLF sorter led to a significant reduction in the overall time it took to obtain a sufficient number of highly pure and viable cells that are suitable for downstream applications like single-cell DNA sequencing. While the WOLF can achieve similar purity alone, the combined technologies reduced enrichment times 15-fold without sacrificing purity. Furthermore, compared to traditional high-pressure cell sorters, this method increases sorting speed without additional shear stress. In summary, this experiment demonstrates a robust workflow for rapidly isolating unique target cell populations without reductions in viability.
APN-012