Methods for Antibody Discovery: Single Cell Techniques in Cell Culture-Based and Microfluidic-Based Technologies
There are many different methods used to perform antibody discovery, including hybridoma, phage display, and single B cell isolation. Here, we will show some examples of the different ways scientists are using single cell techniques to find and develop monoclonal antibodies.
We can break these down into two main focus areas: cell culture-based and microfluidics-based technologies. Each method has its own pros and cons – some methods are expensive and high-throughput, while others are cost-effective but can take a long time.
Hybridomas Method
The most commonly used method of screening B cells is using hybridomas. These were invented in 1975 by Georges J.F. Köhler and César Milstein, who won a Nobel Prize in Physiology or Medicine in 1984 for their efforts. Hybridomas are made by fusing cancer cells with B cells in order to increase the lifespan of the normally short-lived B cells (Figure 1). They are grown in culture and produce monoclonal antibodies (mAbs), so called because they are identical to each other. The hybridoma development process is quite lengthy: it takes approximately 4-8 months to produce purified mAbs that can then be used as diagnostics, therapeutics, prophylactics, and as tools to sort cells with the WOLF®, to give a few examples.
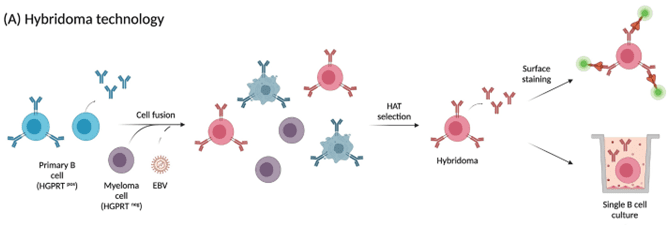
Figure 1: Hybridoma development process. Figure reproduced from Pedrioli and Oxenius, Trends in Immunology 2021. 421143-1158. DOI: 10.1016/j.it.2021.10.008
The hybridoma development process also has a very low throughput with only approximately 1 of 100,000 cells successfully undergoing the fusion process to become an antibody-secreting cell (ASC). This is one reason why it can take so long to go from immunization to ASC. Scientists have made some improvements to the development process by optimizing cell culture conditions for primary B cells and by plating them individually in 96- or 384-well plates. This enabled the screening of a much higher number of cells and thus the discovery of broadly neutralizing antibodies against viruses like HIV or SARS-CoV-2.
Single Cell Plating Method
Single-cell plating can be accomplished by limiting dilution or with a fluorescence-activated cell sorter like the WOLF G2 Cell Sorter with the N1 Single-Cell Dispenser Accessory, which uses microfluidics to gently sort and dispense individual B cells into plates. Once the cells are isolated, there are multiple ways to get to monoclonal antibodies. One way is by using next-generation sequencing (NGS) to obtain the variable heavy (VH) and light (VL) chain gene sequences that can then be synthetically generated or cloned to make the mAbs. (See the application note “Enrichment of Memory B cells for Antibody Discovery and Next-Generation Sequencing” for an example of how the WOLF can be used to do similar experiments.)
A recent preprint from scientists at the Seattle Children’s Research Institute showed a way to use microfluidics to link IgG secretion with surface marker expression and the transcriptional profiles of the ASCs. Using nanovials from Partillion Bioscience that were loaded with single B cells isolated from human donors, the researchers used the WOLF to sort for memory B cells that were secreting IgG and performed single-cell RNA sequencing (scRNAseq) with the 10x Genomics Chromium system (Figure 2).
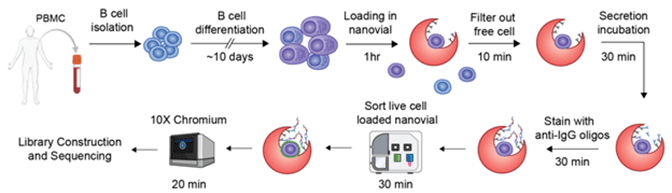
Figure 2: Workflow of SEC-seq. Figure reproduced from Cheng et al. bioRxiv 2022. https://doi.org/10.1101/2022.08.25.505190
They were able to find gene signatures that were associated with high antibody secretion, and hypothesize that this method could be used to better engineer cell secretory function and therefore unlock new therapeutic modalities of antibodies. This could also be applied to shortening the amount of time to screen hybridomas or B cells.
Taking the NGS approach one step further, scientists from Vanderbilt University published a method called LIBRA-seq that linked antibody specificity with the heavy and light chain sequences using scRNAseq. Using antigen baits labeled with both oligonucleotides and fluorophores, they sorted PBMCs for B cells that were able to recognize those antigens and performed scRNAseq using the 10x Genomics Chromium system (Figure 3).
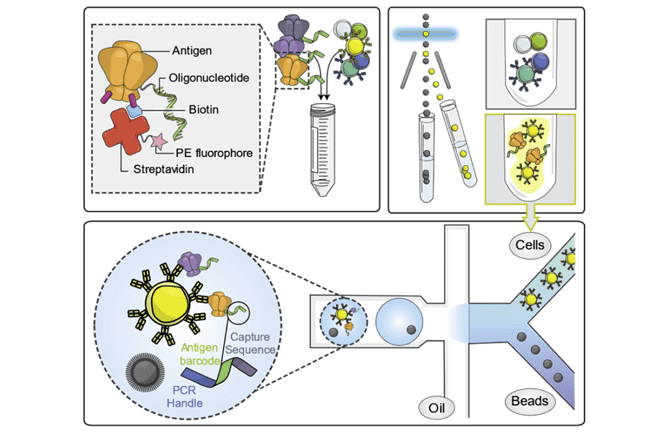
Figure 3: LIBRA-seq workflow. Figure reproduced from Setliff et al. Cell 2019. https://doi.org/10.1016/j.cell.2019.11.003
By linking the unique oligonucleotide sequence of each of the antigens with the unique oligonucleotide sequence on each B cell, they were able to identify a broadly neutralizing antibody against influenza and HIV, as well as predict antigen reactivity of thousands of individual B cells. The researchers made antibodies using the sequence of the heavy and light chains and biologically confirmed the ability of the antibody to bind to influenza virus antigen. In a follow-up publication, the scientists used the same method to identify antibodies against SARS-CoV-2, as did a different group from China.
The Power of Antibody Discovery
The COVID-19 pandemic highlighted the need for a quick, high-throughput way of identifying and mass-producing antibodies for diagnostic, therapeutic, and prophylactic use. We use antibodies to tell us whether we have COVID. Physicians use antibodies to treat COVID and sometimes give them prophylactically to patients who are immunocompromised. Beyond COVID, scientists and physicians have been using antibodies to study or treat autoimmune diseases and cancer for many years. Coming up with better or faster ways to discover new antibodies will help advance medicine and science and improve the lives of humans and animals.
Do you need a better way of performing antibody discovery? Contact us!
Related links:
- https://nanocellect.com/scientific-content/better-cell-sorting-and-antibody-discovery-from-single-bovine-b-cells/
- https://nanocellect.com/scientific-content/enrichment-of-memory-b-cells-for-antibody-discovery-and-next-generation-sequencing/
- https://nanocellect.com/scientific-content/isolate-antibody-secreting-cho-cells-for-single-cell-cloning-with-the-wolf/
- https://nanocellect.com/scientific-content/cloning-bovine-immunoglobulin-vh-and-vl-genes-from-single-b-cells/



