Cell Sorting in Earth’s Microbial Frontiers: A Journey into a Deep Sea Hypersaline Anoxic Basin

In the depths of the ocean lies a mysterious realm known as the deep sea hypersaline anoxic basin (DHAB). These natural geologic basins harbor conditions marked by extreme pressure, intense salinity, and a lack of oxygen, creating unique ecosystems that push the boundaries of life.
In a recent interview, scientist Emily R. Paris, a fourth-year PhD candidate from the Dekas Lab at Stanford University, took us into the world of the ocean’s deepest depths where microbial mysteries await.
Research at Environmental Extremes
As an undergraduate at the University of California, San Diego, Emily became interested in research on microbes in extreme environments after her experiences doing ocean cruises for research expeditions, including her first experience in an extreme environment on a trip to Norway. “I was totally hooked,” she said, reflecting on that early experience.
After completing her bachelor’s degree, she joined the Dekas Lab at Stanford University, which focuses on characterizing the role that bacteria and archaea found in diverse environments play in global nutrient cycles. These environments offer insights into climate change, as well as what factors allow life to persist even in harsh conditions on Earth and potentially on other planets. Emily’s PhD thesis research explores which microbes survive in the very salty conditions of different field sites and how those microbes contribute to their environment.

The Dekas Lab with the ROV onboard the ship (R/V Pt Sur) before setting sail from Louisiana. Left to right: Dr. Anne Dekas (PI), Dr. Steffen Buessecker (Post-doctoral scholar), Emily Paris (PhD candidate).
Microbial Life in the Depths
One of the sites Emily traveled to for her research was the Gulf of Mexico—home to one of the few known deep sea hypersaline anoxic basins (DHABs) in the world—to study the microbes found there. DHABs are natural geologic basins deep beneath the ocean’s surface with water significantly denser and up to ten times saltier than the water above it, resembling massive lakes along the ocean floor.
The extreme environments of DHABs apply several selective pressures to microbes, resulting in a unique microbial population unlike those found in other parts of the ocean. To survive, microbes in DHABs must be able to maintain equilibrium in a high salinity environment, a process that requires a significant amount of energy. Additionally, they must use a metabolism that does not require oxygen due to the anaerobic environment. One of Emily’s research goals is to deepen the base understanding of DHABs and how their unique composition might influence their surrounding environment. Her research addresses some of biology’s most fundamental questions – what defines life and what environmental components are critical for life to survive and thrive.
Using a Niskin Rosette sampling system, Emily and her team were able to capture samples from the anoxic region of the Orca Basin in the Gulf of Mexico. Equipped with sensors, this system recorded the oxygen and salinity levels around it to ensure they were capturing samples from the regions of interest, and enabled analysis of samples across various depths.

Oceans Across Space and Time (OAST) graduate students sampling brine from the Niskin bottles. Going clockwise from top: Chad Pozarycki, Jordan McKaig, Carley Ross, Claire Elbon, Miguel Desmarais, Emily Paris.
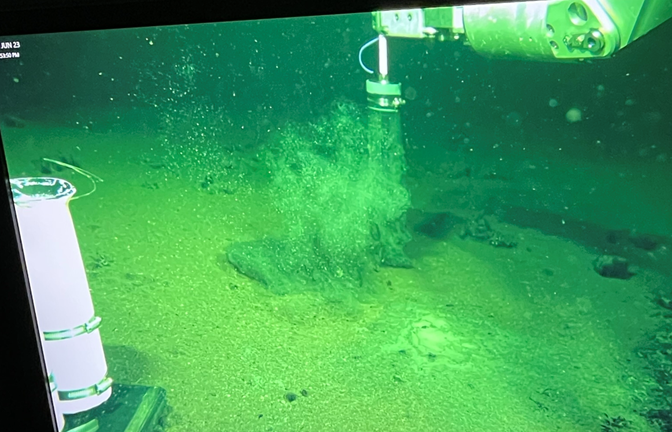
Screenshot from the ROV control room. Here you can see the ROV arm collecting a sediment core from deep within Orca Basin (>2400 m depth). The engineering team had to add tens of pounds to the vehicle to get it to sink into the brine because the high salt content made the vehicle too buoyant to pass through the upper layers on its own. If they added too much weight, the ROV would have sunk into the Jello-like sediment at the bottom of the basin, which would have made vehicle recovery nearly impossible!
A Cell Sorter at Sea
Due to Emily’s collaboration with the Bowman lab at Scripps Institute of Oceanography (the lab that previously took the WOLF Cell Sorter to Antarctica) she embarked on her expedition to the Gulf of Mexico along with Dr. Jeff Bowman who brought the WOLF Cell Sorter on board. The WOLF was stored in a cold room at 4 degrees Celsius to mimic the temperature of the deep sea while sorting and analyzing microbes. Though Emily hadn’t initially planned to incorporate the WOLF into her research, she quickly learned how to use it, and noted that the WOLF had the unique capacity to run high salinity samples with much higher success than other flow cytometers she had previously tested. If she did encounter an issue, she appreciated the flexibility to replace the microfluidic cartridge and try again without risk of damaging the instrument.
For microbial samples from the DHABs, Emily and her team developed assays to detect anabolic and catabolic metabolic activities. Utilizing a stain called redox sensor green, the WOLF allowed Emily to analyze microbial catabolic activity and bulk-sort live, metabolically active microbes based on staining. This approach, combined with metagenome analysis, provided insights into microbial metabolic pathways and enabled comparative analysis between active and inactive cells.

Inside the cold van where the WOLF Cell Sorter was housed (left).
Emily highlighted how the WOLF enabled her to quickly get a readout from the samples they collected while still on the boat. Unlike bulky flow cytometers, the instrument was not only compact enough to travel on the field expedition, but it was user friendly enough for someone with limited flow cytometry experience to quickly learn how to operate and gather meaningful results.
“With my regular research I don’t get results on board; it can take years to get the data. With the WOLF I could get real-time results on board.” –Emily R. Paris
Studying Conditions for Life on Earth and Beyond
Emily’s research has implications for both climate change and astrobiology. With the expansion of oxygen minimum zones in the ocean due to climate change, understanding how microbial populations change and adapt can provide insights into how carbon capture and nutrient cycles may shift in the future. Studying microbial populations in DHABs offers insights into the kinds of microbes that may thrive in low-oxygen conditions and the impact this will have on climate change.
Additionally, Emily’s research contributes to the Oceans Across Space and Time project, funded by NASA’s Astrobiology program. DHABs are thought to be analogs for the oceans found on the icy moons of Jupiter and Saturn – Enceladus and Europa. These moons are believed to have ice oceans in addition to hydrothermal activity along different parts of the sea floor, and they are extremely salty. By characterizing microbial life in DHABs, Emily’s research is helping elucidate the environmental requirements for life and the kinds of metabolisms needed to survive in these extreme, high salinity environments.
Emily’s research sheds light into Earth’s extremes and its climate future and extends to the potential of life in other parts of our solar system.
———
Thank you to Emily for sharing your story and for your passion to pursue research projects that further the understanding of life on our planet and beyond. You can follow the research of Emily and others in the Dekas Lab at: https://dekaslab.stanford.edu/.
Who knew the WOLF would become an important device in Oceanography?
Contact us – let’s see how cell sorting can empower your scientific discoveries, one cell at a time.