Better Cell Sorting and Antibody Discovery from Single Bovine B-Cells
Introduction
A distinct feature of bovine antibodies, ultralong CDR H3 regions, have not been observed in any other species and have the capacity to access epitopes on structurally complex antigens. As such, bovine immunoglobulins have great potential for development as clinical treatments and research tools targeting a broad variety of antigens. However, unlike other species, techniques for studying bovine immunoglobulin genes at the single-cell level have not been comprehensively developed. The Smider group at the Scripps Research and the Applied Biomedical Science Institute established a new method for amplification of bovine immunoglobulin VH and VL genes at the single-cell level utilizing novel cell sorting technology to sort individual B cells into wells for cDNA production and single cell (sc) PCR amplification, followed by nested PCR. Due to high cell integrity, approximately 53% of the wells yielded sequences for both heavy and light chains after sorting and single-cell deposition by the WOLF® Cell Sorter and N1 Single Cell Dispenser.
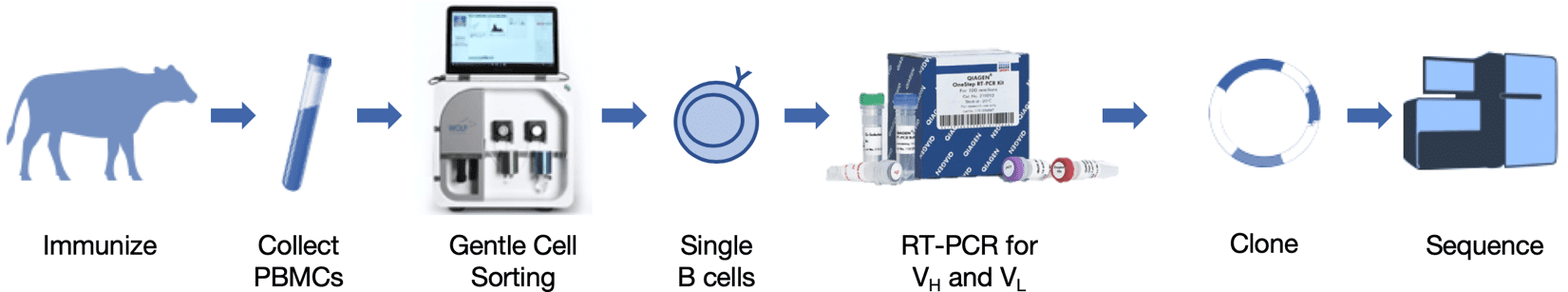
Figure 1. Schematic of heavy and light chain gene amplification from a single bovine B cell. After immunization of a cow with an antigen of interest, blood was collected, PBMCs isolated, and sorted for IgG+ B cells. QIAGEN OneStep RT-PCR Kit, with primers that anneal to mRNA for bovine VH and VL leader peptides and constant regions, was used to produce VH and VL cDNA via specific reverse transcription. VH and VL genes were subsequently amplified with HotStarTaq PCR, then further amplified with nested primers containing restriction sites for cloning. Nested primer PCR products were cloned into expression vectors and sequenced.
Low-pressure sorting improves RNA integrity
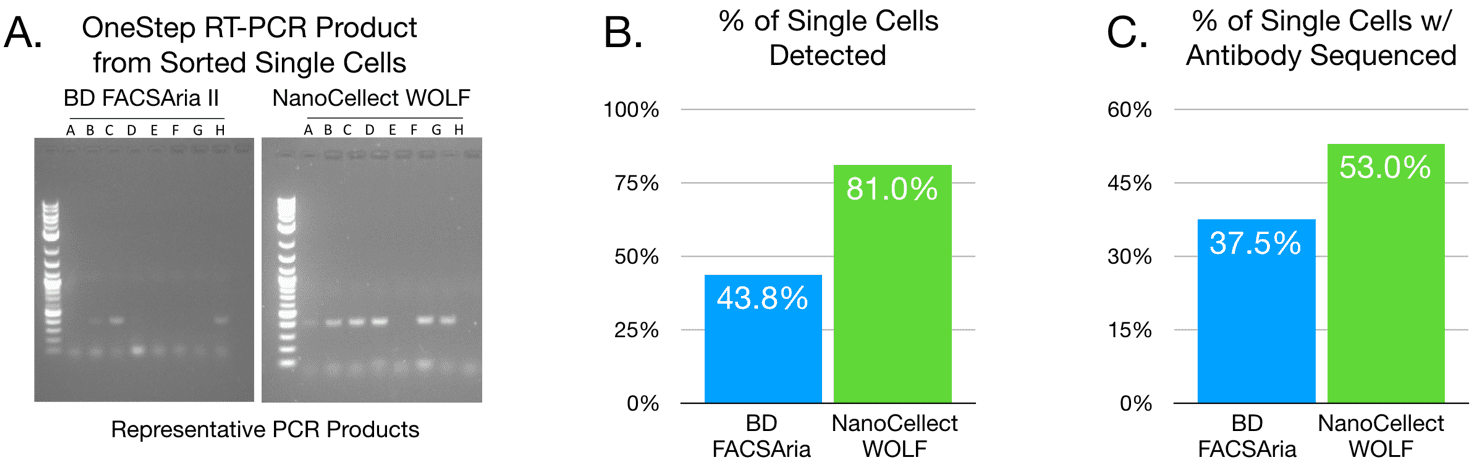
Conventional single-cell sorting into 96-well dispenser plates for scPCR has used complex instrumentation optimized for speed rather than cell viability. Results from individual cells collected on a traditional droplet-based cell sorter (BD FACSAriaTM II) and NanoCellect WOLF Cell Sorter were compared. For PCR amplification of the bovine IgG genes, primers were designed and ratios of each VH and VL primer, along with other conditions, were optimized for production and amplification of cDNA using a QIAGEN OneStep RT-PCR Kit.
Results and conclusion
This is the first protocol to comprehensively optimize amplification of antibody genes from single bovine B cells. Sorting with the WOLF Cell Sorter resulted in nearly 200% more single cells with detectable OneStep RT-PCR product and 41% more single cells were sequenced for both heavy and light chains, compared to traditional FACS. Therefore, the best condition for scPCR to amplify bovine VH and VL chain genes from sorted B cells is highly dependent on the type of cell-sorting instrument used and the optimization of PCR amplification conditions. This new technique’s production of monoclonal antibodies with heavy and light chain pairs expressed from individual bovine B cells will be useful for rapid identification of recombinant bovine antibodies. Little is known about bovine antibody gene repertoires and this method will enable the evaluation of bovine heavy and light chain genetic repertoires at the single-cell level.
Figure 2. The cell sorter impacts scPCR success. (A) Results obtained from cells sorted by BD FACSAria II versus NanoCellect WOLF Cell Sorter were compared for detection of scPCR gel product. (B) Analyzed and combined results (n = 16 for Aria and n = 21 for WOLF) of scPCR product detected. C. Proportion of total number of cells sorted that yielded full antibody sequences (GeneWiz). These results show that percent of cells yielding visible scPCR products and full antibody sequences are higher when cells are sorted by the NanoCellect WOLF Cell Sorter than by BD FACSAria II.
APN-004