The WOLF Outperforms Traditional Sorters in Isolating Primary Hepatocytes for Evaluation of Ploidy Effect on Proliferation and Organoid Outgrowth
Introduction
As the predominant cell type in the liver, hepatocytes perform several metabolic and regulatory processes. To understand liver physiology and disease development, primary hepatocytes can be isolated and cultured to form 3-dimensional organoids that mimic some in vivo liver behavior. However, isolating primary hepatocytes can be challenging.
Traditional fluorescence activated sorters can be too harsh for these fragile, polygon-shaped cells and can lead to substantial drops in viability after sorting. Traditional sorters may also cause cell culture contamination due to their difficult-to-sterilize fluidics and workflow. However, the WOLF® Cell Sorter addresses these issues with a disposable sterile microfluidic workflow that operates at low (<2 psi) pressure to maintain sterility and high viability.
In the study below, the Wang Lab isolated primary hepatocytes to evaluate the effect of ploidy on proliferation. Cells were isolated with both the WOLF and a traditional sorter to determine which offered better organoid formation and outgrowth for future ploidy studies. Ploidy – the quantity of chromosome sets that a cell contains (N) – increases with aging and can inhibit the rate of cell replication in vivo1. Hepatocyte ploidy levels vary substantially so determining their impact on proliferation could improve sorting methods for organoid models and increase the understanding of hepatic physiology.
Method
Cell Preparation
Hepatocytes were isolated from 2-4 month old female mice using a two-step perfusion method with 2.3U of TM Liberase, Enzyme Buffer Solution, and EGTA Buffer2. Non-parenchymal cells (NPCs) were eliminated from the supernatant after a low speed centrifugation. Cell concentrations were adjusted to 2×105 cells/mL of PBS/0.5% BSA and incubated for 10 minutes at 4˚C with Fc block (ThermoFisher, #14-0161-82). Vybrant DyeCycle Green was added according to the manufacturer protocol (ThermoFisher, #V35004), cells were divided into separate aliquots, and immediately sorted on either the WOLF or the BD FACSAriaTM II.
Sorting Based on Ploidy
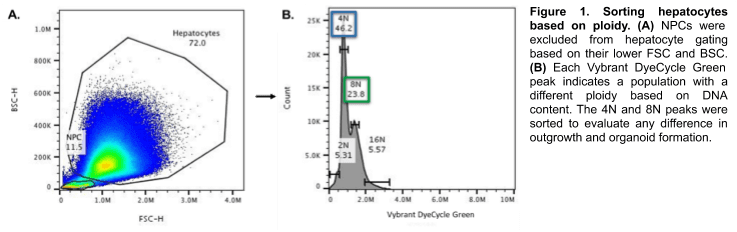
Hepatocytes were gated based on known size and granularity; any remaining NPCs were excluded (Figure 1A). Differing ploidy (DNA content) was identified based on green fluorescence intensity from Vybrant DyeCycle Green (Figure 1B). The 4N and 8N populations were sorted separately into hepatocyte media3.
Post-Sort and Outgrowth Analysis
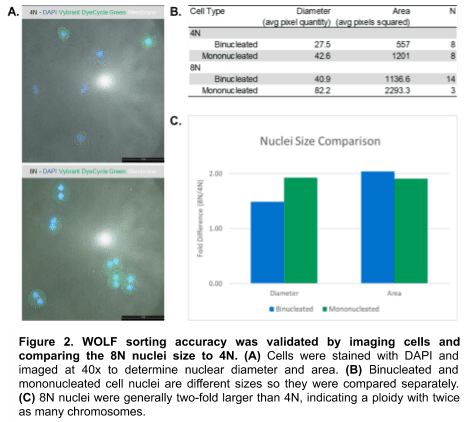
Sorted cells were centrifuged, resuspended in 100 µL of liver wash media3, and counted. 10 µL of each ploidy population was stained with DAPI and imaged using Leica™ software to confirm sorting accuracy (Figure 2A). Each population was split into binucleated and mononucleated sub-populations (which differ in nuclei size) and measured for cell nuclei diameter and area (Figure 2B).
Remaining hepatocytes were centrifuged and embedded as pellets in Matrigel domes. The domes were stored at 37˚C, 5% CO2 in 48-well plates. Media was changed every three to four days and cells were passaged approximately every three weeks. Wells were imaged at Passage 0 and Passage 1 to compare health and proliferation based on sorter and ploidy.
Results
Hepatocyte Nuclei Measurements Indicate Accurate WOLF Sorting
Post-sort ploidy revealed that the 8N hepatocyte nuclear diameters and areas were generally two-folder larger the 4N hepatocytes. This confirms that the WOLF properly sorted and separated 4N and 8N cells (Figure 2).
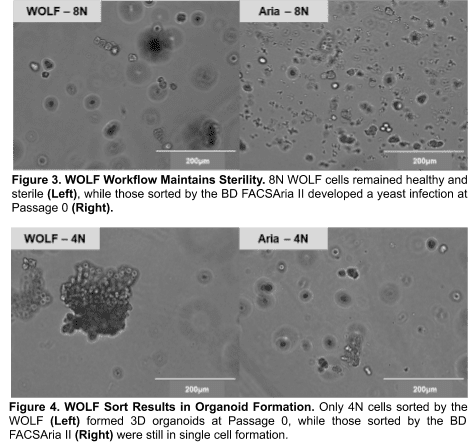

BD FACSAria II Sorted Cells Display Contamination
The 8N population sorted by the BD FACSAria II displayed fungal contamination at Passage 0 in culture and could not be compared to the WOLF for outgrowth (Figure 3). Since all other conditions were shared, contamination was most likely introduced by the BD sorter.
Only WOLF Sort Results in Organoid Formation
At Passage 0, the cells sorted on the BD FACSAria II did not form organoids, likely due to poor recovery from harsh sorting conditions (Figure 4). However, the cells sorted on the WOLF formed 3D organoids with similar morphology as unsorted hepatocyte organoids in previous experiments. Since the WOLF yielded the expected morphology and lacked contamination, it was used for further evaluation of ploidy effect on proliferation.
Higher Ploidy Results in Reduced Hepatocyte Proliferation
The 4N population yielded organoids and higher confluency (Figure 5A) than the 8N population (Figure 5B) at Passage 1. This indicates that the higher 8N ploidy cells proliferate more slowly.
Conclusion
Unlike the BD FACSAria II, WOLF-sorted cells yielded 3D organoids with high confluency and sterility.
The WOLF may have avoided contamination because it uses a disposable cartridge and tubing set, so its fluidics were new and sterile. The BD FACSAria II, like most traditional sorters, uses fixed fluidics that are aseptically cleaned but not sterile, so the contamination could have been introduced there. Another possibility is that the contamination came from the open air during the sort, which could also be avoided with the WOLF – it fits into a biosafety cabinet with HEPA-filtered air.
The BD FACSAria II sorted cells may be sparse and lack organoid formation due to multiple instrument factors that affect viability: higher sorting pressures (20-70 psi), shear forces applied through the sorting nozzle, and decompression shock experienced after exiting the system. The WOLF may have better outgrowth due to its low pressure (<2 psi) sorting mechanism, low shear stress laminar flow, and lack of decompression shock.
The WOLF sorting accuracy was confirmed by 4N and 8N cell nuclei measurements, providing confidence for comparing the two populations. Though the lower fold-difference in binucleated average diameter suggests impure 8N-sorted cells, the two-fold difference in area suggests this may be partially due to manual measurement error instead of sorting impurity. The 8N cells exhibited reduced outgrowth and 3D organoid formation, when compared to the 4N cells, indicating that higher ploidy hepatocytes proliferate more slowly. This confirms previous findings with mouse hepatocytes1.
Here, the WOLF’s sterile, high-viability sorting provided a new platform for studying hepatocyte function. However, these sorting advantages can apply to other aspects of liver cell research, as well as many other sensitive cell types.
References
- Wilkinson, Patrick D., et al. “The Polyploid State Restricts Hepatocyte Proliferation and Liver Regeneration in Mice.” Hepatology, vol. 69, no. 3, 2019, pp. 1242–1258., doi:10.1002/hep.30286.
- Ben-Moshe, Shani, et al. “Spatial Sorting Enables Comprehensive Characterization of Liver Zonation.” Nature Metabolism, vol. 1, no. 9, 2019, pp. 899–911., doi:10.1038/s42255-019-0109-9.
- Peng, Weng Chuan, et al. “Inflammatory Cytokine TNFα Promotes the Long-Term Expansion of Primary Hepatocytes in 3D Culture.” Cell, Cell Press, 29 Nov. 2018, www.sciencedirect.com/science/article/pii/S0
APN-014