Facilitation of Tumorsphere Formation Assay by Isolation of Cancer Stem Cell Markers and Enhanced Single Cell Sorting
Introduction
According to the cancer stem cell hypothesis, tumors are driven and maintained by a small population of cells that display stem-like properties. Researchers have relied on tumorsphere forming assays to try to identify and propagate these cancer stem cells.1 A tumorsphere is a solid, spherical cell formation that develops from a cancer stem cell and is not composed of aggregated cells of other origins. The cells grow in serum-free, non-adherent conditions and can be useful to characterize a cancer stem cell population in vitro.2 This assay can be used with various cell lines such as B16-F10, HT-29, MDA-MB-231,
HCT-116, SKBR3, and MCF-7.
MCF-7 is a model breast cancer research cell line used for mammosphere assays that have cancer stem cell properties detected with markers such as CD44 and CD24. A key step for mammosphere development is to isolate single cells and then calculate the sphere forming efficiency (SFE).3
% SFE= Number of Spheres Counted per Well / Number of Seeded Cells *100
Traditional methods consist of using cell seeding densities ranging from 1,000 to 100,000 cells/mL leading to aggregation of cells, as opposed to only outgrowth from stem cells.4,5 By increasing the number of seeded cells, the sphere forming efficiency decreases. Because mammospheres need to arise from a single cell, plating a single cell per well would be ideal. As a reflection of this, spheroids formed with lower cell densities produce consistent and reliable results when performing drug toxicity studies.6
By using the WOLF G2 Cell Sorter and the N1 Single Cell Dispenser, users can obtain cancer stem cells efficiently by isolating cells with markers of interest to increase sphere forming efficiency by depositing a relevant single cell per well.
Method
Cell and Buffer Preparation
MCF-7 (ATCC HTB-22) cells were grown in T75 flasks with complete growth medium composed of DMEM (Genesee Scientific #25-501), 10% FBS (Genesee Scientific #25-514H), and 1% antibiotic-antimycotic (Invitrogen #15240062).
On the day of sorting, MCF-7 cells were prepared as a single cell suspension using 0.05% trypsin-EDTA (Invitrogen #25300054) and resuspended in sheath buffer in preparation for staining and sorting. The sheath buffer was composed of 1X PBS (Genesee Scientific #25-507), 0.5% BSA (Invitrogen #37525), 5mM EDTA (Invitrogen #AM9260G), and 12.5 mM HEPES (Invitrogen #15630080). The samples and the sheath buffer were kept at 4°C.
MCF-7 cells and SpectraComp compensation beads (Slingshot Biosciences #SSB-05-A) were stained with PE Mouse Anti- Human CD24 (BD Biosciences, #555428) and FITC Mouse Anti-Human CD44 (BD Biosciences, #555478). Prior to staining the cells, MCF-7 cells were blocked with Human TruStain FcX-FC Receptor Blocking Solution (BioLegend #422302) for 10 minutes and then stained with the antibodies. DRAQ7 (Invitrogen #D15107) was used as a viability dye to exclude dead cells from the sort.
Single-Cell Sort and Result Analysis
During analysis, the compensation beads were used to create a compensation matrix to correct for fluorescent channel spillover and single-stained controls were used to set the appropriate gates to sort from a population that expresses CD44high/CD24low (Figure 1).
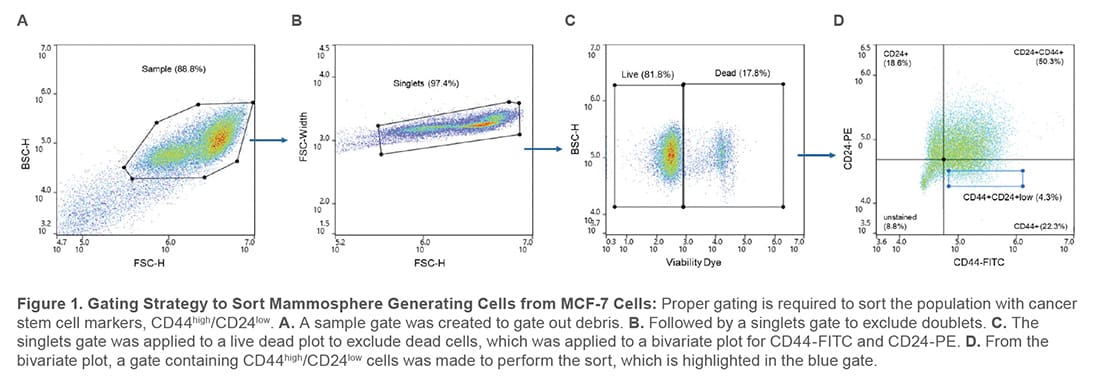
MCF-7 cells were sorted at a concentration of 1 x105 cells/mL using the WOLF and deposited 1 cell/well into 96-well ultralow attachment plates (Crystalgen Inc #522-28032) that were pre-filled with 200 μL of complete cloning medium composed of MammoCult Human Medium Kit (StemCell Technologies #05620) supplemented with 4 μg/mL Heparin (StemCell Technologies #07980) and 0.48 μg/mL Hydrocortisone (StemCell Technologies #07925). A total of three 96-well plates were used to perform the single-cell sort. After sorting, the plates were centrifuged at 100 x g for 3 minutes before incubation at 37°C and 5% CO2.
As a control, an additional single-cell cartridge was used to deposit 10 cells/well to replicate the current techniques. The cells were deposited onto three 96-well plates that were prefilled with 200 μL of the cloning medium, which were then centrifuged at 100 x g for 3 minutes before incubation at 37°C and 5% CO2.
The plates were visualized on Day 0, Day 4, and Day 7 with a change of medium on Day 4 by removing 100 μL of the spent medium and replenishing it with 100 μL of fresh cloning medium. Visualization of the plates was performed to trace the growth of the deposited cells using phase contrast at 10X magnification (ECHO Revolve). Additionally, the spheres were stained with 5 μM of Calcein AM (Invitrogen #C1430) and 25 μg/mL of propidium iodide (PI) (Invitrogen #P3566), which were visualized at 10X magnification using the FITC and TRITC filter cubes (ECHO Revolve).
To determine the spheres with monoclonal origin, the number of mammospheres per well was manually counted on Day 7 and compared to the number of cells deposited on Day 0 to find the mammospheres that originated from a single cell. This number was divided by the total number of wells in the plate. To determine the SFE, the plates prepared at 1 cell/well and 10 cells/well were used to calculate and compare this value. The experiment was replicated in triplicate.
Results
Microscopy Analysis
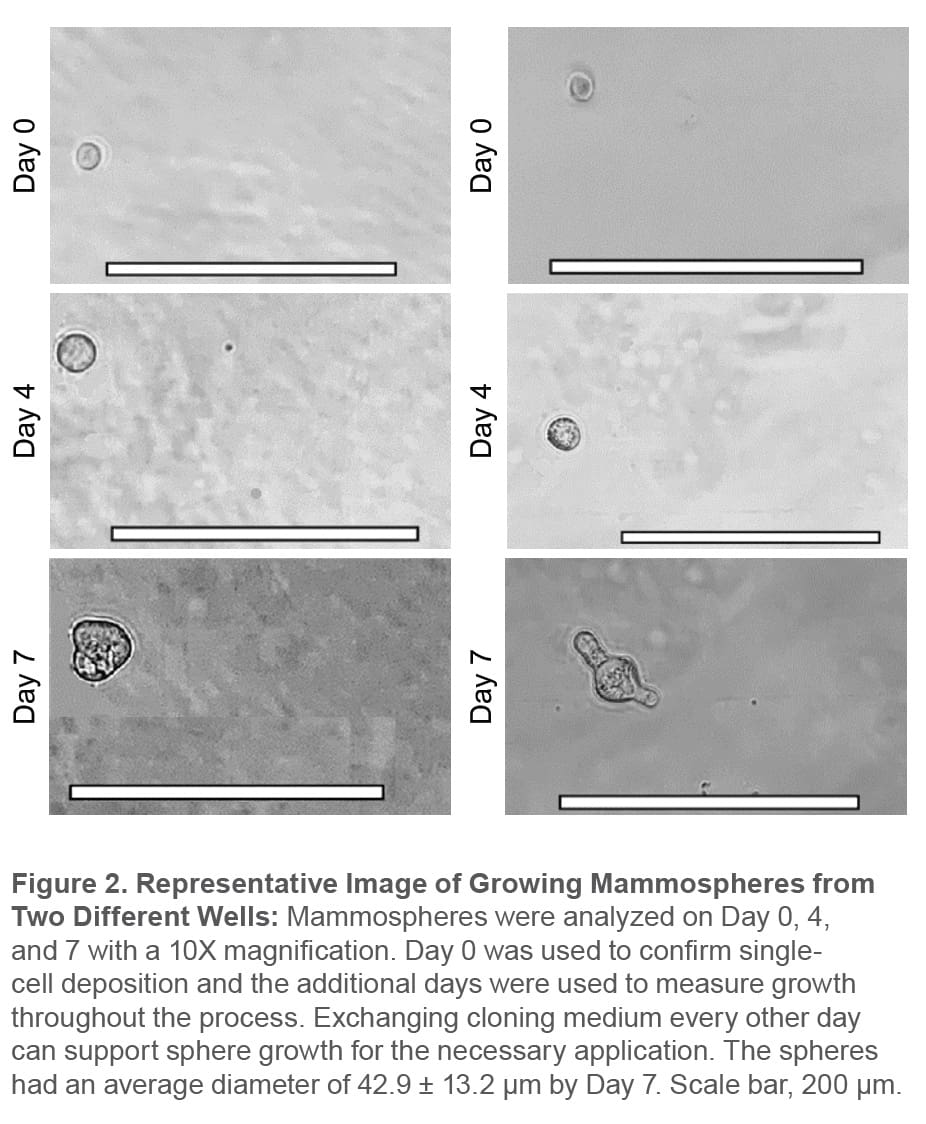
Mammospheres should be solid, round structures with varying sizes from approximately 50 μm to 250 μm.2 This characteristic was confirmed by microscopy. On average, spheres sorted at 1 cell/well on the WOLF had a diameter of 42.9 ± 13.2 μm by Day 7 and the morphology was observed by visualizing growth on Day 0, Day 3, and Day 7 (Figure 2).
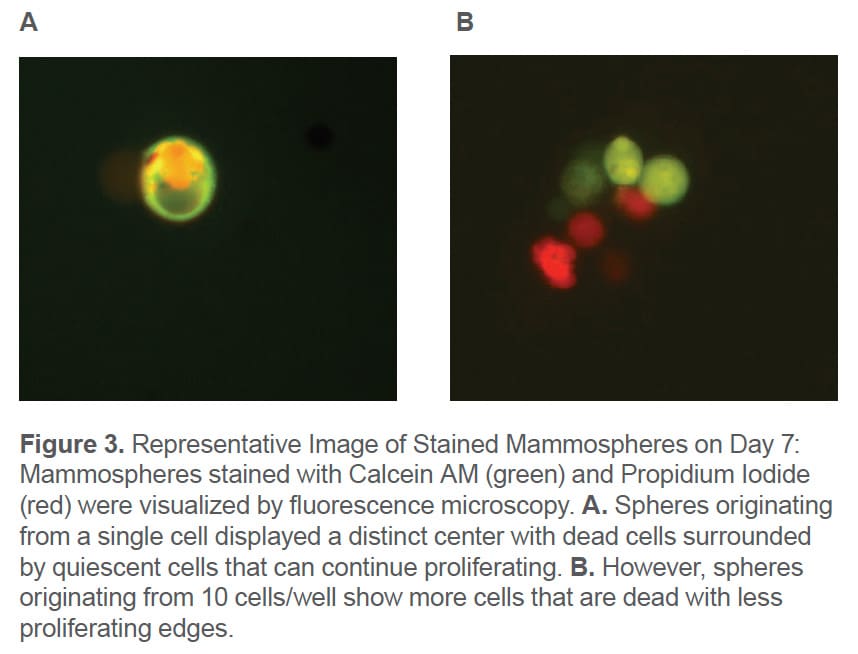
The spheres on Day 7 were stained with Calcein AM to visualize viable cells and PI to demonstrate dead cells. Plating cells at a lower cell density results in less PI staining in the core region (Figure 3A) compared to seeding at 10 cells/well where there are more regions with dead cells (Figure 3B).
In addition, it can be seen that mammospheres prepared at 10 cells/well have a grape-like morphology that is a characteristic of spheroids composed of aggregated cells instead of singlecells. Sphere Deposition and Sphere Formation Efficiency
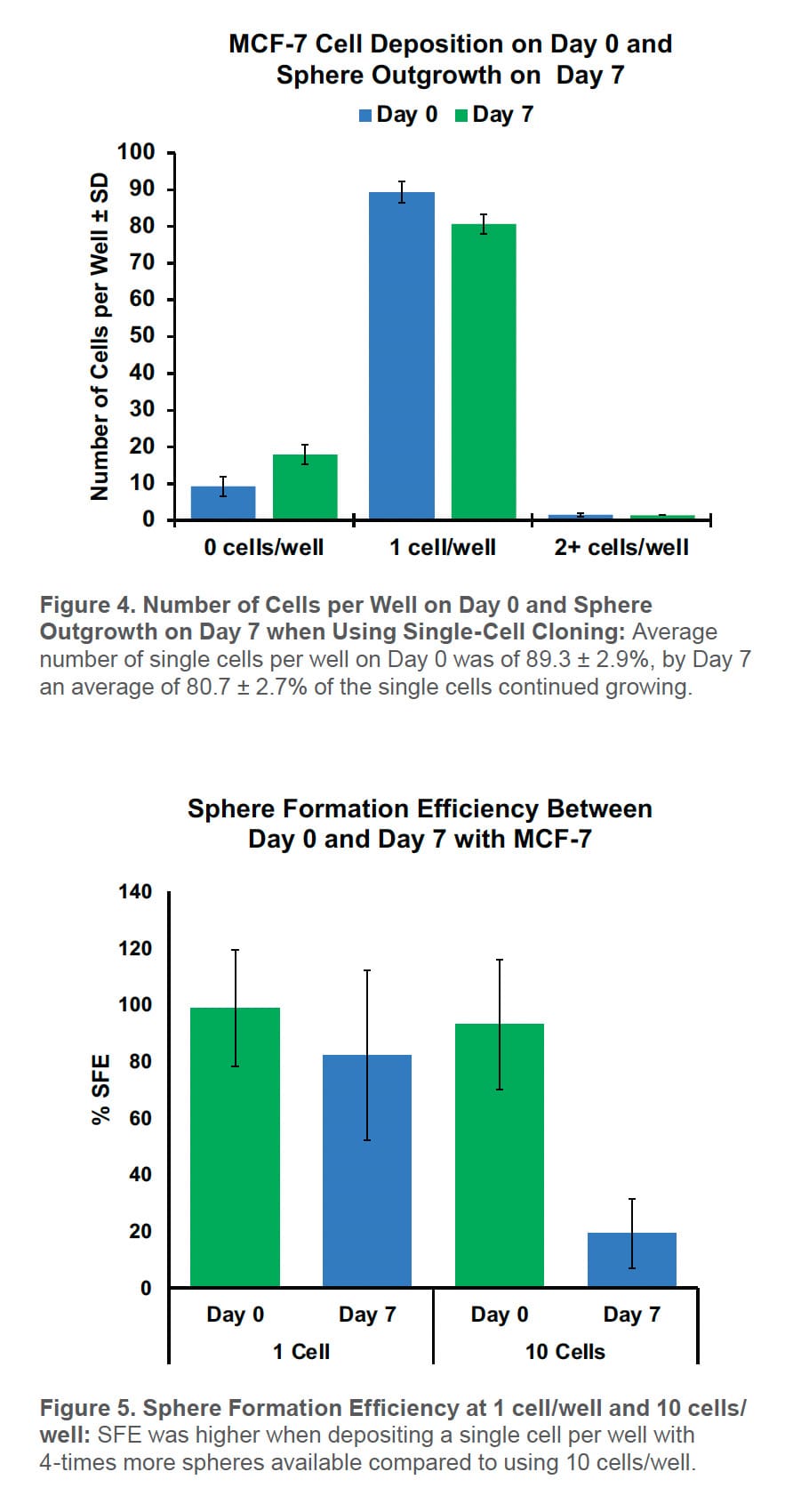
Using the WOLF G2 to sort CD44high/CD24low cells for the mammosphere assay resulted in only 9.2 ± 2.8% empty wells and 1.5 ± 0.4% of the wells contained two or more cells per well on Day 0. On average, 89.3 ± 2.9% of the wells had a single cell. By Day 7, 82.1 ± 2.7% of the wells contained a mammosphere with 80.7 ± 2.7% of them developing from a single cell (Figure 4).
By SFE, there were 4X more mammospheres at 1 cell/well than 10 cells/well on Day 0 (Figure 5). This increases the number of starting spheres needed for downstream applications.
Conclusion
The WOLF G2 platform can be beneficial to support immunooncology and drug screening platforms for mammosphere assays that are useful to further cancer research. In this work, the WOLF G2 demonstrated its efficiency in increasing the number of spheres available per 96-well plate with enhanced monoclonality due to its ability to isolate specific target populations and gently sort them.
For more information, visit nanocellect.com or email [email protected]
References
1. Ospina-Munoz N, Vernot J (2020). Partial Acquisition of Stemness Properties in Tumorspheres Obtained From Interleukin-8-Treated MCF-7 Cells. Sage
Journals, 42(12). https://journals.sagepub.com/doi/full/10.1177/1010428320979438
2. Johnson S, Chen H, Lo P.K. (2013). In vitro Tumorsphere Formation Assays. Bio-protocol, 3(3), e325. https://doi.org/10.21769/bioprotoc.325
3. Bahmad HF, Cheaito K, Chalhoub RM, Hadadeh O, Monzer A, Ballout F, El-Hajj A, Mukherki D, Liu Y, Daoud G, Abou-Kheir W (2018) Sphere-Formation
Assay: Three-Dimensional in vitro Culturing of Prostate Cancer Stem/Progenitor Sphere-Forming Cells. Front. Oncol. 8:347. https://doi.org/10.3389/
fonc.2018.00347
4. Zhu Z, Chen L, Liu J, Huang J, Wu G, Zheng Y, Yao K (2017). A Novel Three-Dimensional Tumorsphere Culture System for the Efficient and Low-Cost
Enrichment of Cancer Stem Cells with Natural Polymers. Exp Ther Med 15: 85-92. https://doi.org/10.3892/etm.2017.5419
5. Bailey, P. C., Lee, R. M., Vitolo, M. I., Pratt, S. J., Ory, E., Chakrabarti, K., Lee, C. J., Thompson, K. N., & Martin, S. S. (2018). Single-Cell Tracking of Breast
Cancer Cells Enables Prediction of Sphere Formation from Early Cell Divisions. IScience, 8, 29–39. https://doi.org/10.1016/j.isci.2018.08.015
6. Singh, S. K., Abbas, S., Saxena, A. K., Tiwari, S., Sharma, L. K., & Tiwari, M. (2020). Critical role of three-dimensional tumorsphere size on experimental
outcome. BioTechniques, 69(5), 333–338. https://doi.org/10.2144/btn-2020-0081
APN-036