Enrichment of Memory B Cells for Antibody Discovery and Next-Generation Sequencing
Introduction
Hybridoma technology has made it possible to obtain monoclonal antibodies and has become an essential tool for developing therapeutic drugs that can combat diseases such as HIV, Ebola, and, most recently, SARS-CoV-2.1 However, there are well known limitations of the technology, including under sampling the natural diversity of the antibody repertoire and genetic drift of the cell line over time.2 A way to bypass these limitations is to use B cells directly to discover human monoclonal antibodies. This process consists of screening and isolating antigen-specific B cells from lymph nodes, spleen, or whole blood. The B cells are then cloned to generate higher yields of antibody for further in vitro and in vivo assays.3
Memory B cells are highly sought for human antibody discovery because of their in vivo development and affinity maturation, resulting in antibodies that have high specificity and affinity towards target proteins.4 They are distinguished from naive CD19+ B cells by the expression of CD27, a cell-surface marker. Long-lived memory B cells in peripheral blood express surface IgG, or less frequently IgM, compared to naïve B cells, which mostly produce IgM when a primary antigen is encountered to later develop into IgG.5,6 Sequencing the antibody repertoires of both isotypes shows the diversity of B cell lineages and somatic hypermutation levels, indicating diversity of antigen specificities and affinity maturation, respectively.7
Using the WOLF Cell sorter, we enriched a population of CD27+ B cells. This process was validated using Abterra Bioscience’s Reptor next-generation sequencing (NGS) service. Reptor is a flexible and fast sequencing service that provides accurate antibody repertoire analysis for any researcher. The WOLF combined with Reptor empowers any lab to rapidly interrogate the repertoires of antigen-specific memory B cells.
Method
Whole Blood Preparation
Whole blood from a healthy donor was mixed 1:1 with sample buffer composed of 1X HBSS (Gibco 14175-079) and 2% FBS (GenClone 25-514H). Diluted blood was then added onto a 50 mL SepMate Tube (STEMCELL Technologies 85450) that was prefilled with Lymphopure (STEMCELL Technologies 07801), which was followed by centrifugation at 1200 x g for 10 minutes. The sample was then decanted into a new tube containing sample buffer and centrifuged at 350 x g for 5 minutes with low brake. The supernatant was removed and the cells were resuspended in FBS with 10% DMSO (Sigma D8418) and stored at -80°C until sorting.
Cell Staining
The stored sample was thawed and washed with sample buffer. The cells were then blocked with 10% FBS. After blocking, the cells were stained with an antibody mixture composed of FITC anti-human CD3/CD8a/CD14 (BioLegend 300406, 301006, 367115), and PE-Cy5 anti-human CD27 (Invitrogen 15027941). Finally, the cells were stained with SYTOX Green Ready Flow (Invitrogen R37168). The sample was then diluted to a final concentration of 1.0 x 106 cells/mL using sample buffer.
Bulk Sorting for Memory B Cell Enrichment
Live CD3-CD8-CD14-CD27+ cells were sorted into collection tubes containing FBS. The waste tube (dump channel) which contained CD3+CD8+CD14+CD27- cells, was also saved for downstream sequencing. The post-sort sample was analyzed on the WOLF to determine post-sort purity. This post-sort sample and the sample from the dump channel were also used for further sequencing of IgG and IgM.
Sequencing of the Bulk Sorted Cells
For sequencing analysis, 2,500 sorted live CD3-CD8-CD14-CD27+ cells and 2,500 of the unsorted CD3+CD8+CD14+CD27- cells from the dump channel were lysed in 200 µL of lysis buffer (QIAGEN 79216). The total RNA was isolated and eluted in 20 µL of Zymo Quick RNA MicroPrep Kit (Zymo R1050). cDNA was synthesized in 10 µL reactions using oligo (dT) primers. Nested PCR was used to amplify the complete variable domain of both IgM and IgG cDNA. The amplicons were then indexed with Nextera XT adapters (Illumina), and loaded onto a MiSeq instrument (Illumina) to generate 300bp x 300bp paired end reads. Each amplicon library was sequenced to a depth of 500K reads and analyzed with Reptor software (Abterra Biosciences). Briefly, Reptor stitches paired end sequencing reads, corrects sequencing errors, annotates CDR sequences, and organizes the repertoire into clonal lineages based on CDR3 sequence.
Results
Purity of Enriched Memory B Cells Using Bulk Sorting
The WOLF was able to successfully enrich live CD3- CD8-CD14-CD27+ cells from an average pre-sort target population of 2.4 ± 1.2% to a post-sort target of 81.5 ± 0.8% when using a starting sorting concentration of 1.0 x 106 cells/ mL (Figure 1).
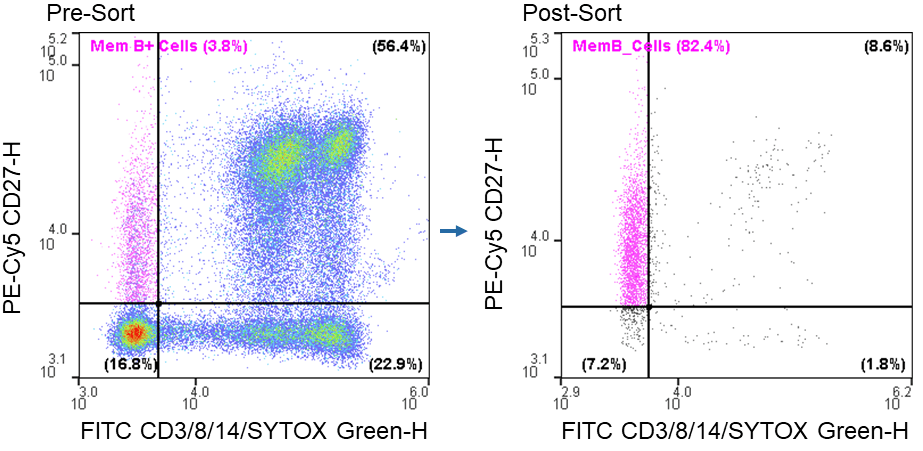
Figure 1. Memory B cell post-sort purity: Sorting of live CD3-CD8-CD14-CD27+ from PBMCs resulted in a 34-fold increase in purity.
Sequencing of Enriched Memory B Cells
The IgG and IgM transcripts of 2,500 CD27+ enriched cells and CD27- cells collected in the dump channel were sequenced and analyzed by Abterra Biosciences.
After repertoire construction, a total of 2,108 unique IgG and IgM sequences were observed in the enriched CD27+ cells. Additionally, the enriched cells indicated higher levels of hypermutation (reflecting affinity maturation) than the CD27- cells (Table 1). The number of distinct CDR3 sequences also matches the expectation that CD27+ cells are antigen-exposed and therefore show focusing of CDR3s as compared to antigen-naïve CD27- IgM cells.
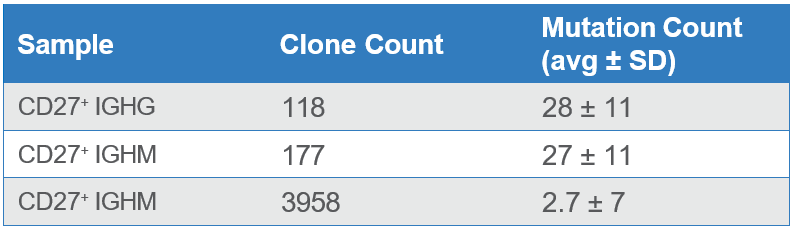
Table 1. Counts of unique clones (CDR3’s) and mutation rates across antibody sequences from enriched CD27+ cells and from CD27- cells from the dump channel.
The higher mutational loads observed in the enriched sample indicate that a majority of the CD27+ memory B cells were purified using the WOLF. Meanwhile, the sample collected in the dump channel displayed lower mutation rates because it contained a higher number of CD27- cells (Figure 2).
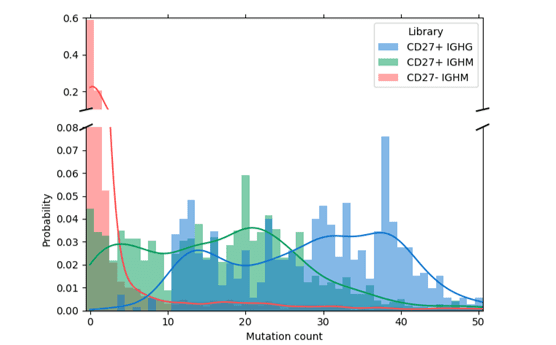
Figure 2. Mutation distribution for IgG and IgM in CD27+ enriched cells: Antibodies with a greater number of mutations have undergone affinity maturation in response to antigen exposure. This is observed in the enriched CD27+ memory cells denoted in green and blue bars where mutation probability is higher. Meanwhile, the sample from the CD27- dump channel (red bars) contained fewer mutations.
Conclusion
In conclusion, when pairing gentle sorting of CD27+ cells with the Reptor NGS analysis, users can further validate that the sorted cells contain a diverse pool of IgG and IgM sequences and show that hundreds of antigen-exposed clones were enriched from the background of naïve B cells. This workflow highlights the value of a gentle, microfluidic cell sorter combined with a B cell sequencing service to improve the efficiency of antibody discovery that can be implemented by any lab.
For more information, visit nanocellect.com and abterrabio.com or email [email protected]
References
1. Lu, R-M., Hwang, Y-C., Liu, I-J., Lee, C-C., Tsai, H-Z., Li, H-J., & Wu, H-C. (2020). Development of therapeutic antibodies for the treatment of diseases. Journal of Biomedical Science, 27(1). https://doi.org/10.1186/s12929-019-0592-z
2. Crowe JE. (2009). Recent advances in the study of human antibody responses to influenza virus using optimized human hybridoma approaches. Vaccine, 27: G47-G51. doi: 10.1016/j.vaccine.2009.10.124
3. Tiller (2011). Single B cell antibody technologies. New biotechnology, 28(5), 453–457. https://doi.org/10.1016/j.nbt.2011.03.014
4. Pedriolo A, Oxenius A (2021). Single B Cell Technologies for Monoclonal Antibody Discovery. Trends in Immunology 42(12). https://doi.org/10.1016/j. 2021.10.008
5. Stewart A, Chi-Fung Ng J, Wallis G, Tsioligka V, Fraternali F, and Dunn-Walters (2021). Single-Cell Transcriptomic Analyses Define Distinct Peripheral B Cell Subsets and Discrete Development Pathways. Front Immunol., 12:602539. doi: 10.3389/fimmu.2021.602539
6. Takemori T (2015). B Cell Memory and Plasma Cell Development. Molecular Biology of B Cells. 227-249. https://doi.org/10.1016/B978-0-12-397933- 00014-X
7. Yaari G, Kleinstein SH. (2015). Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Medicine, 7:121. https://doi.org/10.1186/ s13073-015-0243-2
APN-028
