Enrichment and Growth of Transgenic Microalgae Using the WOLF G2 Cell Sorter
Introduction
Microalgae are a diverse group of organisms that play critical ecological roles as primary producers and contribute substantially to aquatic photosynthesis, which generates ~50 % of the oxygen present in the earth’s biosphere1,2. Microalgae are also increasingly leveraged in biotechnology applications for the production of renewable fuels, food, nutraceuticals, and other bioproducts3–6. Fluorescence activated cell sorting can be used to enrich desired populations of microalgal isolates in fundamental research, such as the discovery and isolation of new species, and in the development of stable engineered strains for various biotechnology applications7.
Chlamydomonas reinhardtii is perhaps the most well-studied microalga and serves as a model for understanding numerous biological processes including photosynthesis, chloroplast biology, circadian systems, carbon-concentrating mechanisms, and environmental stress responses8–11. Moreover, due to its haploid genome and ease of growth in culture, C. reinhardtii is used by scientists as a platform for the development of genetic tools and as a chassis for production of various products10,12. Here we use Chlamydomonas as an archetypal microalga to demonstrate the gentle sorting capabilities of the WOLF G2 ® cell sorter in conjunction with the N1 single cell dispenser. With this platform, GFP positive cells can be highly enriched and maintain high viability after sorting single cells to inoculate 200 µl cultures in 96-well plates. These results indicate the potential for the WOLF G2 ® and N1 Single-Cell Dispenser to transform a diverse array of microalgal workflows, from laboratory strain development to single-cell genomics on a research vessel on the open ocean.
Methods
Solid and Liquid Medium
Liquid: One liter of fresh 1x Tris-Acetate-Phosphate liquid medium was made by combining 3.17g of Tris-Acetate-Phosphate powder (Phytotech Labs, cat# T8224), 1.0 mL glacial acetic acid (Millipore Sigma, cat # A6283), and 1.0 mL of Hutner’s trace elements (https://www.chlamycollection.org/) into 800 mL deionized water stirring in a plastic beaker on a magnetic stir plate until all components were in solution. The pH was adjusted to 7.0 and the solution brought up to a final volume of 1.0 L with deionized water. The solution was then filter-sterilized through a 0.22 µm filter apparatus (Thermo Scientific, cat# 569-0020) in a biosafety cabinet and stored room temperature in low light.
Agar plates: a 2x liquid TAP medium and a warm 3% agar base (Teknova, cat# A1790) were combined to make a 1.2% final agar/1x TAP mixture. Using an electronic pipette aid (Drummond Scientific, cat# 4-000-105) and a 100 mL sterile serological pipet (UX-04395-22), 30 mL of warm solution was poured into each 100×25 mm petri dish (VWR, cat# 89107-632). Agar plates were poured in a biosafety cabinet to maintain the sterility.
Strains
Axenic cultures of Chlamydomonas reinhardtii CC-4533 (cw15 mt- [Jonikas CMJ030])13 and CC-5416 (met1(UVM)-47A mt-) were acquired from the Chlamydomonas resource center (https://www.chlamycollection.org/) at the University of Minnesota, St. Paul, USA. Strain CC-4533 is a cell wall-deficient wild-type background strain and CC-5416 is a nuclear-localized GFP constitutive expressing line.
Culture System and Growth Curves
CC-4533 and CC5416 were grown up independently from streaks on agar slants and inoculated into 50 mL of Tris-Acetate-Phosphate (TAP) liquid medium in 125 mL disposable culture flasks (BioPioneer, cat# DGFPC0125S). Cultures were incubated at 20°C (room temperature) on an orbital shaker moving at 100 rpm under constant low light 20 µE m-2 sec-1 (see Endnote).
Colonies were re-streaked onto fresh 1x TAP plus 1.2% agar plates monthly. Plates streaked from active seed cultures were also created as a back-up. Plates were incubated stationary benchtop, 20°C, under constant low light 20 µE m-2 sec-1.
Growth curves for CC-4533 and CC-5416 were performed to determine growth conditions. New 50 mL cultures were inoculated at a cell density of 3.0×105 cells/mL and placed on an orbital shaker rotating at 100 rpm, 20°C, under constant low light 20 µE m-2 sec-1. Two 300 µL aliquots (technical replicates) from each line were taken daily and analyzed for cell number using the Novocyte flow cytometer (Agilent Technologies) with a 405/488/561 laser configuration. Once cell cultures reached a density of 1×106 cells/mL, aliquots for analysis required a 1:10 dilution to maintain linearity of the data.
Microscope Images
10x and 40x bright field and fluorescence images were taken on an inverted light/fluorescence microscope pre and post sort to confirm cell intactness and enrichment of viable phenotype.
Sorting Chlamydomonas with WOLF G2®
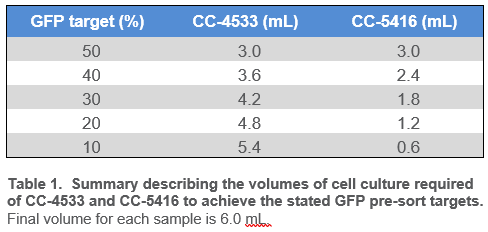
Chlamydomonas CC-4533 and CC-5416 strains were grown to a cell density between 1.0×106 and 3.0×106 cells/mL and were diluted with fresh 1x TAP to a final density of 5.0×105 cells/mL. Cell counts were determined on the Novocyte flow cytometer (Agilent Technologies) with a 405/488/561 laser configuration. Phenotypes for CC-4533 (wild-type) and CC-5416 (constitutively expressing nuclear GFP) were confirmed via microscopy prior to sorting (Figure 5). Cell lines were then combined at various ratios to a final volume of 6.0 mL (Table 1). Each mixed culture was then filtered through a 40 µm nylon sieve to remove clumps and large particles, which could clog the cartridge on WOLF G2.
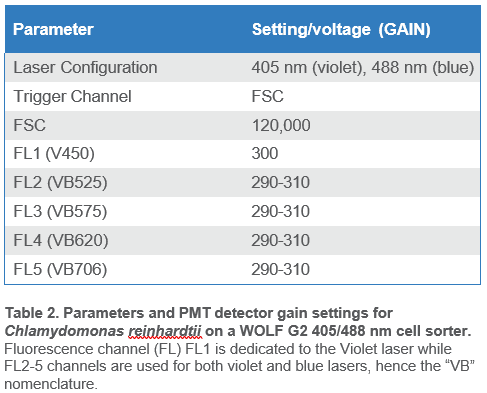
When ready to sort, 3.0 mL of filtered cells were transferred to a sterile 5.0 mL FACS tube. The WOLF was primed and calibrated with sterile 1x TAP medium as sheath. The WOLF N1 single-cell plate dispenser was calibrated alongside WOLF G2 with 1x TAP. Sterile, flat-bottom 96-well plates (Corning, cat# CLS3340) were filled with 200 µl of 1x TAP medium in each well for the single-cell sorting experiments. Sorting parameters are summarized in Table 2.
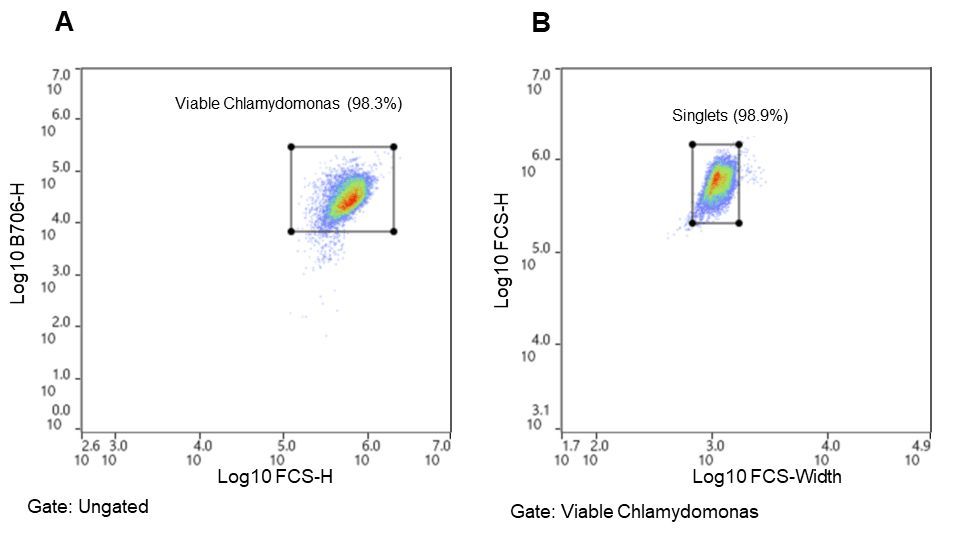
Chlamydomonas cells were identified using chlorophyll autofluorescence and cell size estimated from scatter (Figure 2)8,9,12. The gating strategy was as follows: all events shown above the 104 decade are considered positive for chlorophyll and therefore likely algal cells; this is the parent gate (A). Following the parent gate is a child plot exclusive of singlet events to reduce coincidence in sorting, and it is gated to the parent (B). Third is a sub child plot displaying GFP positive events, which is inclusive to events in the singlets child plot (C). Lastly, (D) is a histogram representing the distribution of GFP positive events in the population. Parameters and voltage gains of the sorts are listed in Table 2.
NOTE: It is suggested to pause the sort and invert the FACS tube gently every 30 minutes for the duration of the sort to keep cells in suspension and to prevent cartridge clogging.
Results
Growth curves were conducted before each experiment to determine how quickly a culture of Chlamydomonas reinhardtii grows from linear to stationary phase under the provided growth conditions (refer to the Methods). Data suggest rapid growth occurs between day 1 and 3 (Figure 1). Therefore, sorting experiments were conducted using culture from the early growth phase.
Pre-sort GFP targets for bulk sorting ranged from 50% to 10% (Table 1), while the pre-sort single-cell GFP target was 10%. Viable Chlamydomonas reinhardtii cells were identified by observing chlorophyll autofluorescence emission (FL4; B620) and GFP emission (FL2; B525); gating strategy is described in Figure 2. Chlorophyll autofluorescence is confirmation of algal or plant-derived events8–10,12. In addition, the strength of the chlorophyll autofluorescence signal is a direct representation of algal cell viability8,9,12. While this signal varies between algal cell species and across lines, a high signal for Chlamydomonas reinhardtii on WOLF G2 indicates viable cells and is typically above the 104 decade while lower signals, below the 104 decade, represent stressed/dying cells.
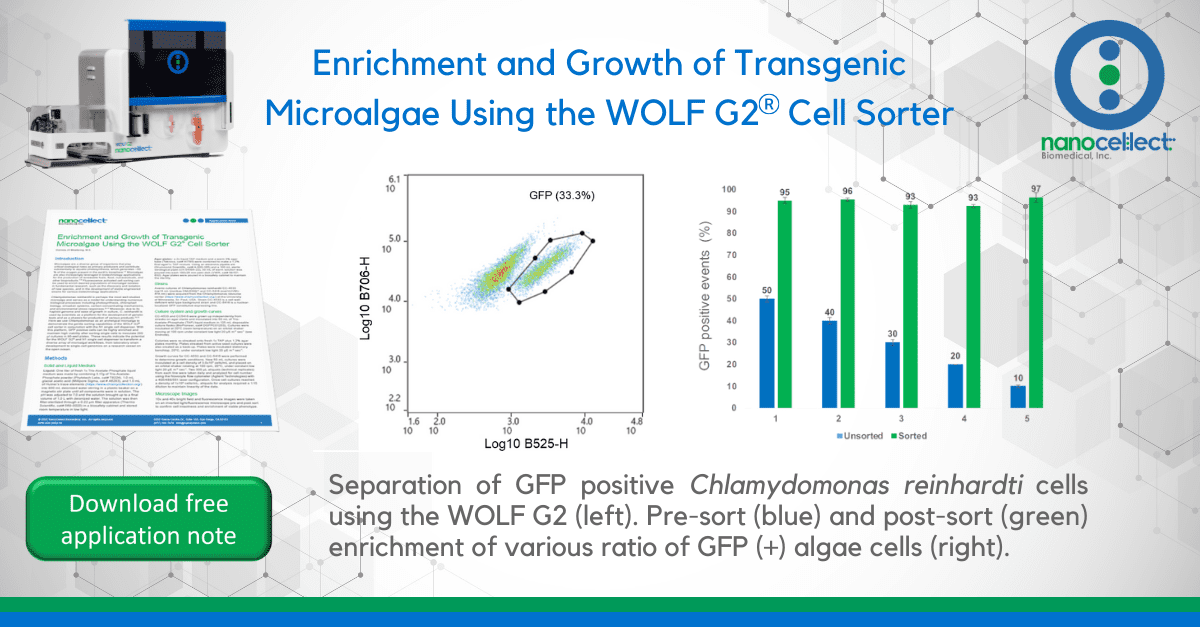
Bulk sorting data show an enrichment of viable GFP positive cells ranging from 93-97% post sort (Figures 3-4). In addition, data also suggest that enrichment of GFP positive cells at 90% or greater on WOLF G2 is achievable regardless of the starting pre-sort target.
Single-cell data suggest positive long-term viability and growth of sorted GFP positive cells three weeks post sort (Figures 6-7). Growth of single cells in 200 µl cultures in 96 well plates ranged from 80-82% across two independent experiments. Phenotype confirmation of single-sorted cells post sort is shown in Figure 7. In three weeks, one sorted cell per well successfully created a clonal population of many cells.
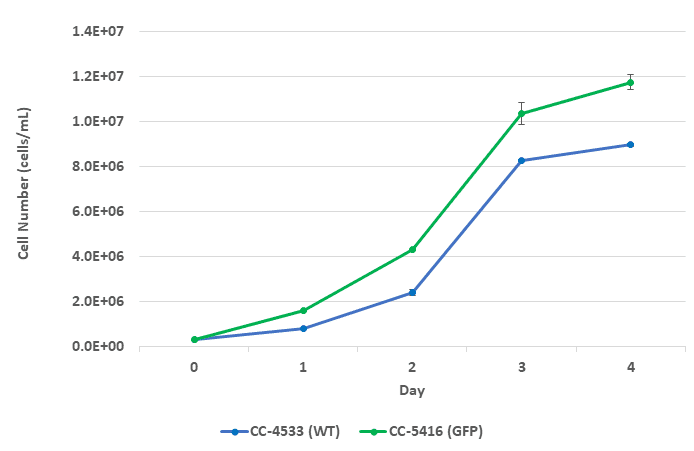
Figure 1. Growth curve for CC-4533 and CC-5416 over a 4-day period. Green-colored data points represent CC-5416 (GFP) and blue-colore -data points represent CC-4533 (wild-type). Error bars represent the standard deviation of N=2 technical replicates.
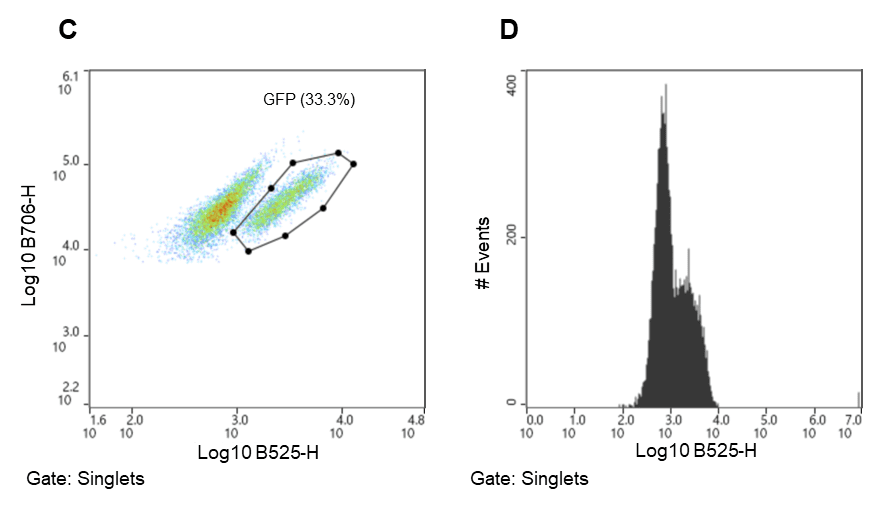
Figure 2. Chlamydomonas reinhardtii sorting strategy. Algal cells were separated using chlorophyll autofluorescence (FL5, B706) and estimated cell size (FSC) both excited by the 488nm laser. A. Parent gate (chlorophyll positive events – viable Chlamydomonas), B. Child gate selecting for singlet events (gated to parent), C. Subsequent child gate selecting for GFP positive events (gated to singlets) (FL2, B525), D. A histogram representation of the GFP positive events within the child gate “singlets.”
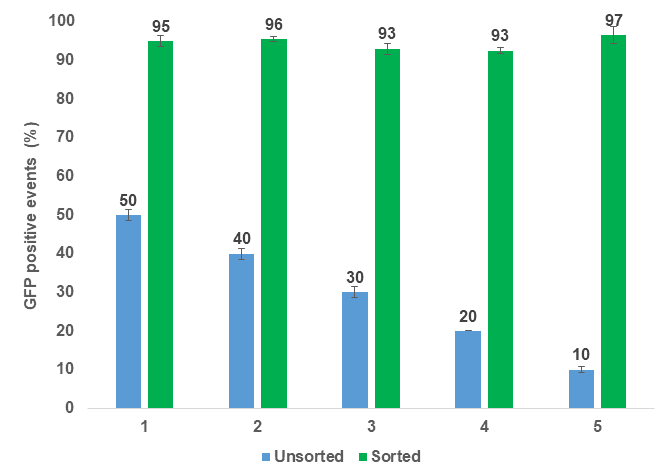
Figure 3. Bulk enrichment of GFP positive Chlamydomonas reinhardtii. Five independent experiments were performed over the span of three weeks. Blue bars represent unsorted algal cells with varying target populations of GFP. Targets were achieved by mixing CC-4533 (WT) and CC-5416 (GFP) cells together at different ratios. Green bars represent enriched/sorted GFP positive cell populations. Error bars represent the standard deviation of N=2 technical replicates.
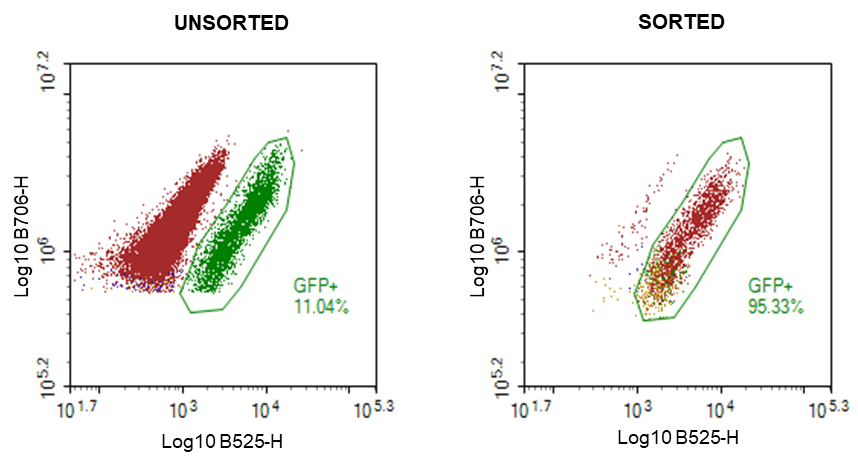
Figure 4. Bulk enrichment confirmation of GFP positive Chlamydomonas reinhardtii using the Novocyte flow cytometer: A. Unsorted Chlamydomonas mixed at a 90:10 ratio (WT CC-4533: GFP CC-5416) , and B. Sorted Chlamydomonas GFP cells. Gating strategy is identical to that used on WOLF G2 405/488 (Figure 2).
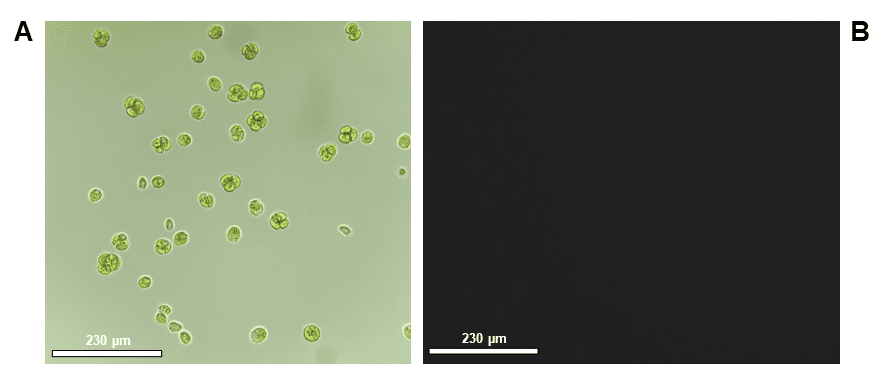
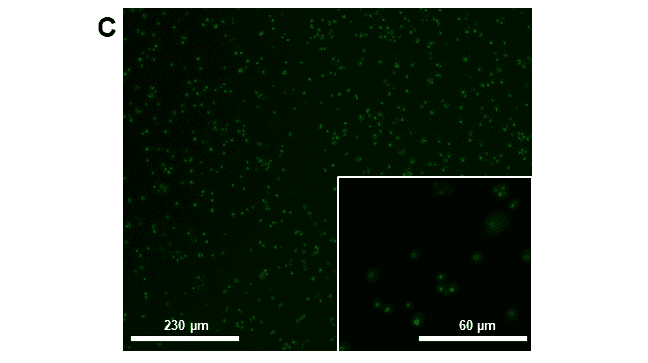
Figure 5. Microscope images taken for confirmation of phenotype pre-sort: A. CC-4533 (WT) bright-field, 40x objective, B. CC-4533 (Wild-type) fluorescence image using FITC cube, 40x objective and C. CC-5416 (GFP), 10x and 40x objectives. Scale bars are representative of relative cell size on the 10x or 40x objectives, 230 µm or 60 µm, respectively. Images were taken on an inverted ECHO microscope. GFP fluorescence exposure time was 150 ms.
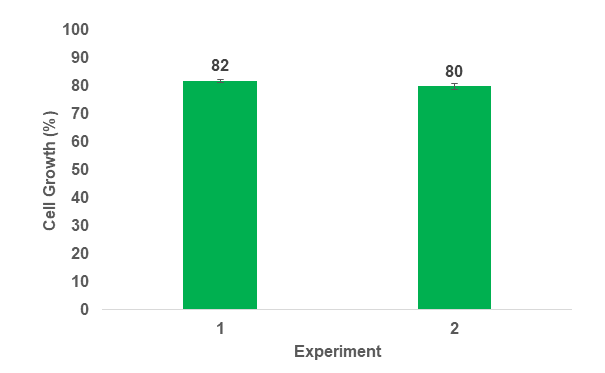
Figure 6. Chlamydomonas reinhardtii single-cell growth 3 weeks post sort. Green bars represent two independent experiments. Error bars represent the standard deviation of N=3 and N=4 replicate plates, respectively.
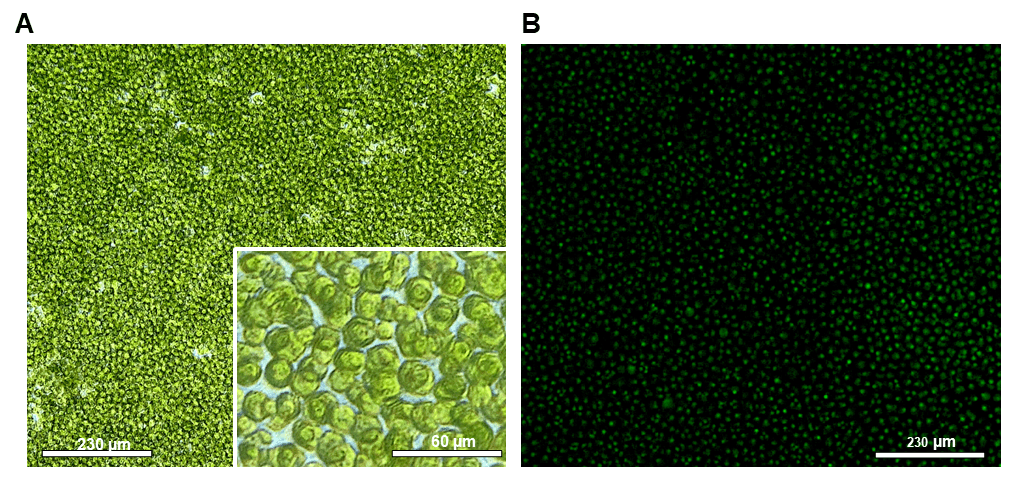
Figure 7. Post sort images of Chlamydomonas reinhardtii, CC-5416 (GFP) using the WOLF N1 single-cell dispenser. Pre-sort GFP target population was 10% CC-5416. Images are representative of a well three weeks post sort. A. 10x and 40x bright-field images of sorted CC-5416, and B. 10x FITC/ GFP fluorescence image. Scale bars are representative of relative cell size on the 10x or 40x objectives, 230 µm or 60 µm, respectively. Images were taken on an inverted ECHO microscope. GFP fluorescence exposure time was 150 ms.
Conclusions
We conducted bulk and single-cell sorting experiments to assess enrichment of viable GFP positive cells using the WOLF G2. WOLF G2 was able to gently enrich for viable GFP positive Chlamydomonas reinhardtii cells, which showed high growth after a three-week incubation. The combination of high enrichment (>93%) and high viability of sorted cells (>80%) provides a platform for scientists to enhance their workflows for a variety of applications including stable cell line development for various bioproducts including biofuels, species/mutant isolation and identification, -omics studies, ocean monitoring and health, and discovery/isolation of new algal species†. The WOLF G2 system is ideal for field work due to its small size, portability, and ease of use. The microfluidics system provides a gentler sorting environment when compared to traditional jet-air sorters and is an ideal choice where algal cell viability is of utmost importance.
For more information, visit nanocellect.com or email [email protected]
Endnote:
Chlamydomonas reinhardtii is a robust and rapidly growing microalgae10,14. To maintain vigorously growing cultures, a new seed culture should be inoculated weekly from fresh colonies on streaked agar plates. This limits the number of subcultures created from a single seed culture and prevents the transfer of potential stress affects to new cultures if, for example, the seed cultures have entered nutrient depletion.
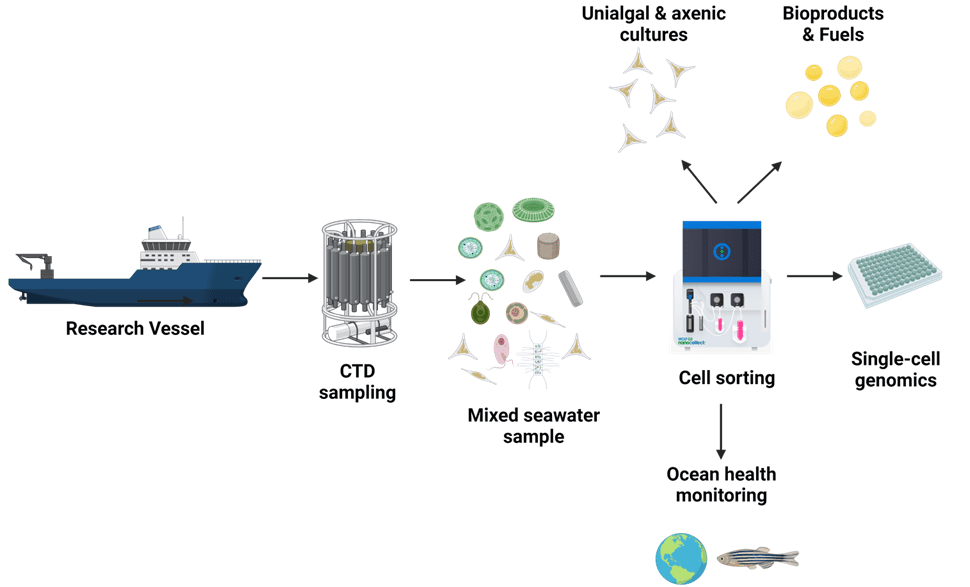
†Diverse applications in microalgal sorting:
Gentle cell sorting of microalgae has a wide range of applications. Enrichment and growth of transgenic stains in bulk and via single-cell dispensing, is a steppingstone for deeper experimentation and discovery to answer global environmental questions. Below is a visual workflow summarizing several possible applications.
References
1. Falkowski, P. The power of plankton. Nature 483, 517–520 (2012).
2. Chapman, R. L. Algae: The world’s most important ‘plants’-an introduction. Mitig Adapt Strateg Glob Chang 18, 5–12 (2013).
3. Xia, D., Qiu, W., Wang, X. & Liu, J. Recent advancements and future perspectives of microalgae-derived pharmaceuticals. Marine Drugs vol. 19 Preprint at https://doi.org/10.3390/md19120703 (2021).
4. Kratzer, R. & Murkovic, M. Food Ingredients and Nutraceuticals from Microalgae: Main Product Classes and Biotechnological Production. (2021) doi:10.3390/foods.
5. Blasio, M. & Balzano, S. Fatty Acids Derivatives From Eukaryotic Microalgae, Pathways and Potential Applications. Frontiers in Microbiology vol. 12 Preprint at https://doi.org/10.3389/fmicb.2021.718933 (2021).
6. Ren, Y., Sun, H., Deng, J., Huang, J. & Chen, F. Carotenoid production from microalgae: Biosynthesis, salinity responses and novel biotechnologies. Marine Drugs vol. 19 Preprint at https://doi.org/10.3390/md19120713 (2021).
7. Schroda, M. & Remacle, C. Molecular Advancements Establishing Chlamydomonas as a Host for Biotechnological Exploitation. Frontiers in Plant Science vol. 13 Preprint at https://doi.org/10.3389/fpls.2022.911483 (2022).
8. Cheloni, G. & Slaveykova, V. I. Morphological plasticity in Chlamydomonas reinhardtii and acclimation to micropollutant stress. Aquatic Toxicology 231, (2021).
9. Terashima, M., Freeman, E. S., Jinkerson, R. E. & Jonikas, M. C. A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid Chlamydomonas mutants. (2014) doi:10.1111/tpj.12682.
10. Salomé, P. A. & Merchant, S. S. A Series of Fortunate Events: Introducing Chlamydomonas as a Reference Organism [OPEN]. doi:10.1105/tpc.18.00952.
11. Fauser, F. et al. Systematic characterization of gene function in the photosynthetic alga Chlamydomonas reinhardtii. doi:10.1038/s41588-022-01052-9.
12. Sproles, A. E., Berndt, A., Fields, F. J. & Mayfield, S. P. Improved high-throughput screening technique to rapidly isolate Chlamydomonas transformants expressing recombinant proteins. Appl Microbiol Biotechnol 1, 3.
13. Kurniasih, S. D. et al. UV-mediated Chlamydomonas mutants with enhanced nuclear transgene expression by disruption of DNA methylation-dependent and independent silencing systems. Plant Mol Biol 92, 629–641 (2016).
14. Gross, M.-M., Zelzion, J., Price, E. C. & Levitan, D. C. Using Natural Selection to Explore the Adaptive Potential of Chlamydomonas reinhardtii. PLoS One 9, 92533 (2014).
APN-038