Efficient Cell Sorting of Microalgae for Biotechnological Innovations
Introduction
Cell sorting of microalgal species is important to support the evolution of fuel alternatives, development of biological medicines, and continued production of food supplements1–3. Potential high-value substances are also further driving the exploration of microalgae as unique biotechnological production systems4–9. Flow cytometry methods are already firmly established in algal environmental and toxicological studies and would be greatly beneficial for the development of biotechnological processes, such as enrichment of cells with high oil phenotypes, or gene-edited cells for improved biologics2,5,6,10. More recently, Arthrospira platensis has been gaining attention as a potential host for biologic drug development because of its high concentration of soluble proteins, which can convert into high expression of therapeutic proteins1,4.
Arthrospira platensis, also known as Spirulina, is a symbiotic, multicellular, and filamentous cyanobacteria (blue-green microalgae), which fixes nitrogen from air and has a rather complex life cycle (Figure 1). This cyanobacterium is the most cultivated photosynthetic prokaryote; its biomass is widely used as a health food, feed supplement, and as a source of fine chemicals1,3. The total annual production of biomass is estimated to be 12,000 metric tons, more than 50% of which is produced in China and the greater Asia-Pacific region1. Utilizing cell sorting for the enrichment of genetically improved Spirulina cells could help speed up sustainable biological drug development.
Two major challenges when sorting algal cells, like Spirulina, on a fluorescence-activated cell sorter are the size/morphology of cells and the behavior of the cells in culture: they can exist as single cells and as colonies (mats), both of which can clog the flow cell on a cell sorter. Mature Spirulina filaments are rather long (200-500+ µm) and skinny (~ 8.0 µm in diameter) and since they are filamentous in nature, they aggregate to form mats1,6. Here we show evidence that Spirulina filaments of various lifecycle stages and lengths can successfully be separated in a gentle manner on the WOLF G2® Cell Sorter and are able to grow into full cultures.
Methods
Strain and Culture Maintenance
An Arthrospira platensis starter kit was ordered from the Algae Research Supply (cat# AKAp-00500). Sterile 1x Spirulina culture media was obtained from the UTEX culture collection of algae, University of Texas at Austin11.
A total of 1.5 mL of starter culture was added to 50 mL of sterile 1x Spirulina liquid media in each of two 125 mL polycarbonate Erlenmeyer shaker culture flasks (TriForest, cat# FPC125S). The flasks were incubated at 20°C on an orbital shaker moving at ~50 rpm under a 12 hr Light:12 hr Dark photoperiod (~60 µE m-2 sec-1). Cultures were serially passaged every two weeks with a 1:10 dilution of culture to medium. After three passages the culture adjusted to its new environment and was ready for experimental use.
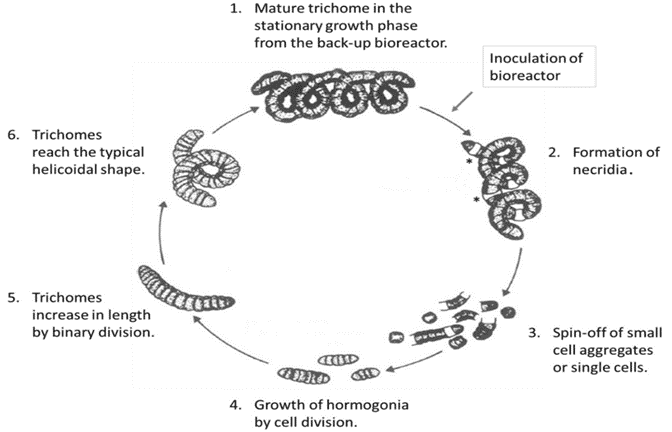
Figure 1. Lifecycle of Arthrospira platensis (Spirulina)12.
Microscope Images
Bright-field images were acquired on an ECHO revolve microscope pre- and post-sort to confirm cell intactness, enrichment of viable phenotype, and growth 1-2 weeks thereafter.
Sorting Spirulina with WOLF G2®
CHO-K1 cells were diluted at a final concentration of 2.0 x 106 cells/mL in PBS/1% BSA (Thermo Scientific 37525). The cells were then stirred at 200 rpm at room temperature using the stirrer function of the CS1 module. The stirrer function was turned on and the sample was analyzed on the WOLF G2 every 15 minutes for one hour to measure if the cell concentration remained consistent while stirring.
Sorting on WOLF® G2
Spirulina was passaged and grown for 1.5 weeks under the conditions stated above. In a laminar flow hood, 3.0 mL of culture was diluted 1:1 with culture medium and then gently sieved through a sterile 70 µm nylon filter (FalconTM, cat# 352350) to remove clumps.
When ready to sort, 3.0 mL of filtered cells were transferred to a sterile 5.0 mL FACS tube (Corning, cat# 352057). The WOLF G2 was primed and calibrated with sterile 1x Spirulina culture medium as sheath.
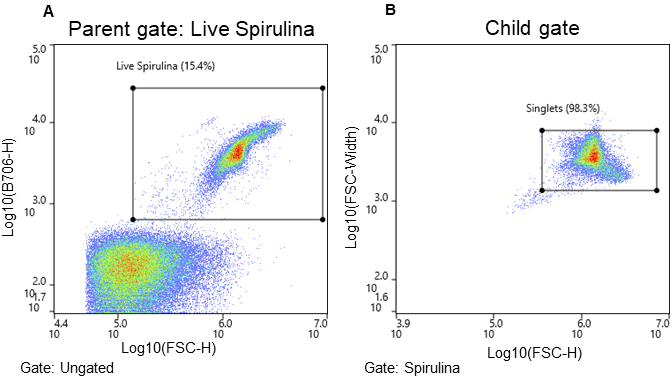
Spirulina cells were identified using chlorophyll autofluorescence, and cell size was estimated from light scatter (Figure 2). The gating strategy was as follows: all events above the 103 decade are considered positive for chlorophyll and therefore likely algal cells; this is the parent gate (2A). Following the parent gate is a child plot exclusive to singlet events to reduce coincidence during sorting, and it is gated to the parent (2B). Third is a sub-child plot displaying three gates labeled “early stage, short chain, and long chain” to represent three potential phenotypes, which are inclusive of events from the singlets child plot (2C). Lastly, (D) is a scatterplot to monitor potential clogs in the cartridge during the sort. Clogs would present themselves as vertical gaps in the data. Parameters and voltage gains of the sort are listed in Table 1.
NOTE: The sort was paused and the FACS tube was gently inverted every 30 minutes for the duration of the sort to keep cells in suspension and prevent cartridge clogging.
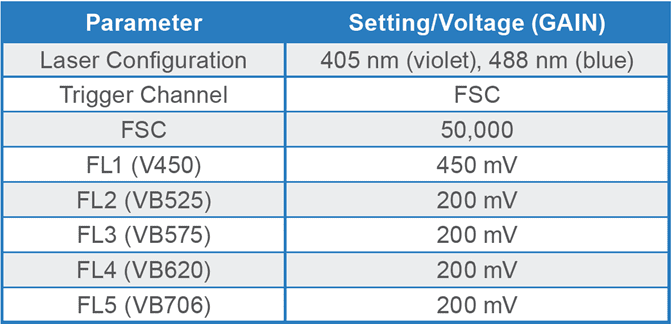
Table 1. Parameters and PMT detector gain settings for Arthrospira platensis on the WOLF G2 405/488 nm cell sorter. Fluorescence channel (FL) FL1 is dedicated to the violet laser while FL2-5 channels are shared between the violet and blue lasers, hence the “VB” nomenclature.
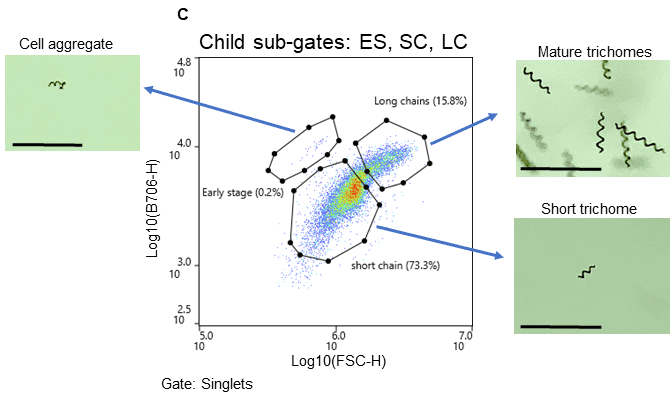
Figure 2. Spirulina sorting strategy and associated microscope images. Cells were separated using chlorophyll autofluorescence (FL5, B706) and estimated cell size (FSC-H) both excited by the 488nm laser. A. Parent gate (chlorophyll positive events – viable Spirulina), B. Child gate selecting for singlet events (gated to the parent), and C. Subsequent child gates selecting for three populations of Spirulina cells (early stage (ES), short chain (SC), and long chain (LC). The bar in images represents 230 µm.
Results
Viable Spirulina cells were identified by observing chlorophyll autofluorescence emission (FL4; B620); the gating strategy is described in Figure 2. Chlorophyll autofluorescence is confirmation of algal or plant-derived events13–15. In addition, the intensity of the chlorophyll autofluorescence signal for Spirulina on the WOLF G2 indicates viable cells and is typically above the 103 decade, while signals below the 103 decade represent stressed/dying cells and debris.
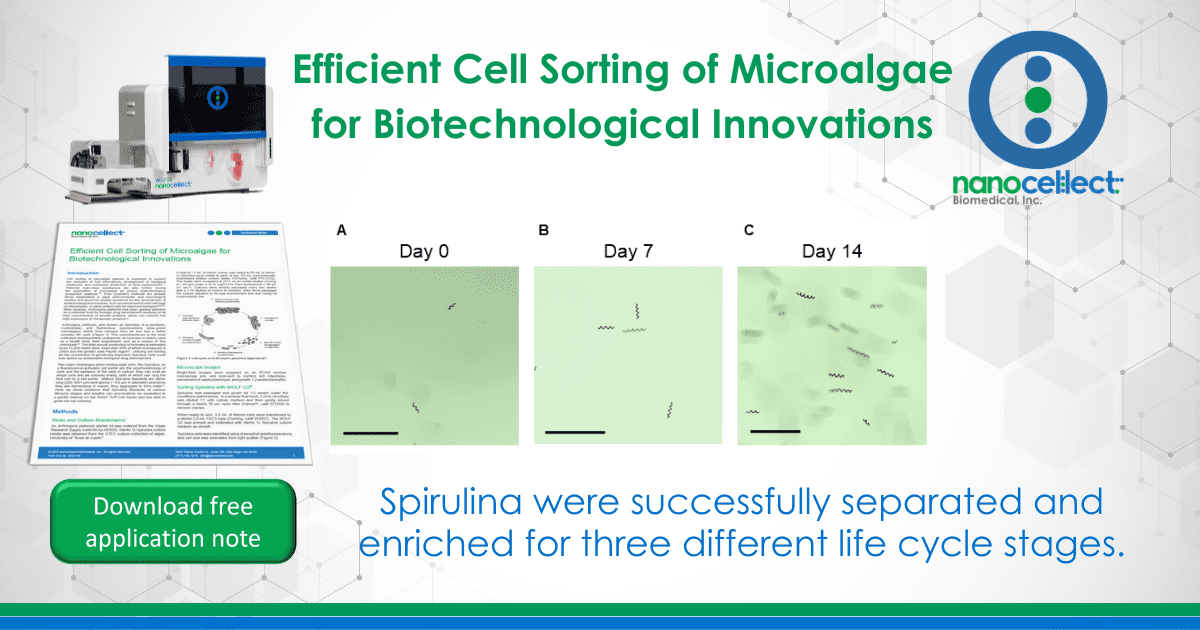
Bulk sorting data show that the WOLF G2 successfully separated and enriched for three different life cycle stages of Spirulina (Figure 2). Short chain (16%) and long chain (73%) Spirulina trichomes were sorted with purity while the “early stage (0.2%)” presented a mixed but smaller population of hormogonia, small cell aggregates, and single cells (Figures 1, 2C). Furthermore, short chain and long chain populations were maintained for 2 weeks post-sort to determine if cells could grow into full cultures, which they did successfully (Figure 3). No cartridge clogging was observed.
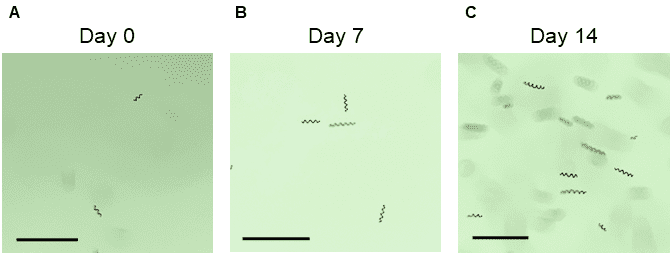
Figure 3. Microscope images of short-chain Spirulina up to two weeks post-sort: A. immediately post-sort (Day 0), B. seven days post-sort, and C. fourteen days post-sort. The bar in all images represents 230 µm.
Conclusion
Spirulina is considered a large, irregular cell type due to its length and helical form and is therefore a challenge to sort through traditional cell sorters. Using the WOLF G2 Cell Sorter, we were able to identify and separate Spirulina with varying chlorophyll levels, representing three different lifecycle stages. These were successfully grown in culture for up to two weeks post-sort, demonstrating that the WOLF G2 did not affect the viability or regeneration capability of Spirulina.
For more information, visit nanocellect.com or email [email protected].
References
1. Masojídek, J. & Torzillo, G. Mass Cultivation of Freshwater Microalgae. Reference Module in Earth Systems and Environmental Sciences (2014)
doi:10.1016/B978-0-12-409548-9.09373-8.
2. Falkowski, P. The power of plankton. Nature 483, 517–520 (2012).
3. Chapman, R. L. Algae: The world’s most important ‘plants’-an introduction. Mitig Adapt Strateg Glob Chang 18, 5–12 (2013).
4. Lumen Bioscience: Biologic drugs reimagined. lumen.bio.
5. Hyka, P., Lickova, S., Přibyl, P., Melzoch, K. & Kovar, K. Flow cytometry for the development of biotechnological processes with microalgae. Biotechnol Adv 31, 2–16 (2013).
6. Singha Roy, A., Bhowmick, D., Pal, R. & Roy Sikhsha Prangan, C. PHYTOPLANKTON COMMUNITY STUDY FROM EUTROPHIC WETLAND EMPLOYING FLUORESCENCE ACTIVATED CELL SORTING METHOD Biodiesel from green algae View project FlowCytometry of Human & Mice Adipoyte View project JOURNAL OF INTERNATIONAL ACADEMIC RESEARCH FOR MULTIDISCIPLINARY PHYTOPLANKTON COMMUNITY STUDY FROM EUTROPHIC WETLAND EMPLOYING FLUORESCENCE ACTIVATED CELL SORTING METHOD. vol. 4 www.jiarm.com (2016).
7. Xia, D., Qiu, W., Wang, X. & Liu, J. Recent advancements and future perspectives of microalgae-derived pharmaceuticals. Marine Drugs vol. 19 Preprint at https://doi.org/10.3390/md19120703 (2021).
8. Kratzer, R. & Murkovic, M. Food Ingredients and Nutraceuticals from Microalgae: Main Product Classes and Biotechnological Production. (2021)
TCN-013