Intact Nuclei Sample Preparation from Frozen Brain Tissue with the WOLF Cell Sorter
Download PDF
Introduction
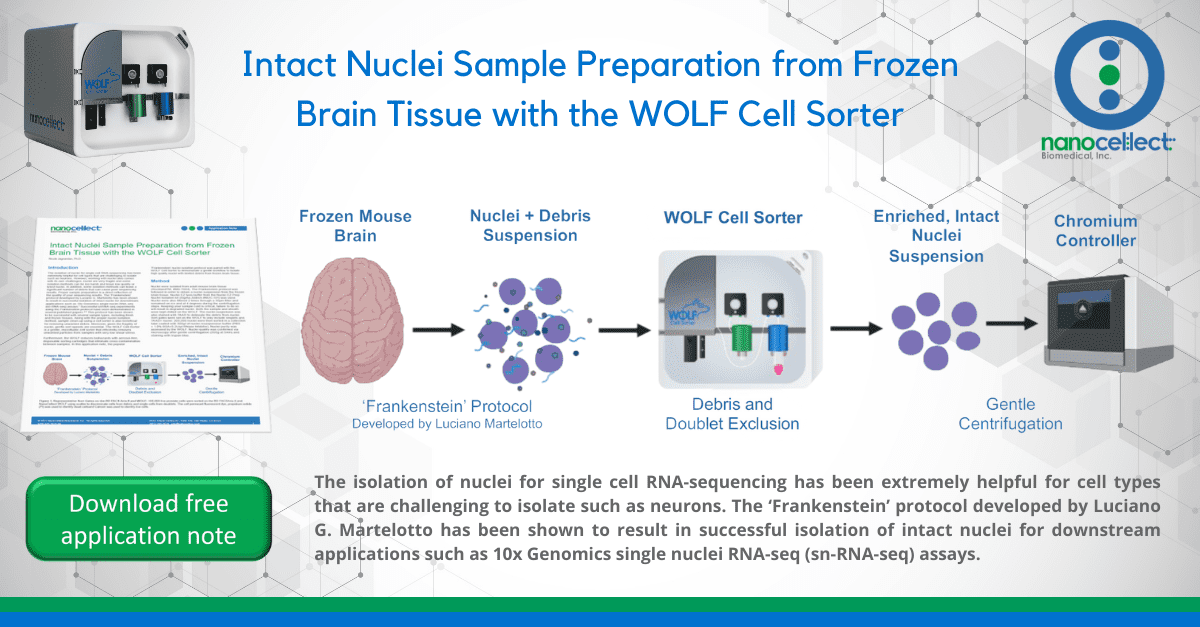
The isolation of nuclei for single cell RNA-sequencing has been extremely helpful for cell types that are challenging to isolate such as neurons. However, working with nuclei also comes with its own challenges, nuclei are very fragile and some isolation methods can be too harsh and leave low quality or lysed nuclei. In addition, some isolation methods can leave a significant number of debris that can cause poor sequencing results. Proper sample preparation is a direct reflection of the quality of your sequencing results. The ‘Frankenstein’ protocol developed by Luciano G. Martelotto has been shown to result in successful isolation of intact nuclei for downstream applications such as 10x Genomics single nuclei RNA-seq (sn-RNA-seq) assays.1 Successful snRNA-seq experiments using the Frankenstein protocol have been demonstrated in several published papers.2-5 This protocol has been shown to be successful with several sample types, including fresh and frozen tissues. Along with the proper nuclei isolation method, sample clean-up using a cell sorter is also beneficial for removing unwanted debris. Moreover, given the fragility of nuclei, gentle sort speeds are essential. The WOLF Cell Sorter is a gentle, microfluidic cell sorter that efficiently removes unwanted particles from samples with very low shear stress.
Furthermore, the WOLF reduces biohazards with aerosol-free, disposable sorting cartridges that eliminate cross contamination between samples. In this application note, the popular ‘Frankenstein’ nuclei isolation protocol was paired with the WOLF Cell Sorter to demonstrate a gentle workflow to isolate high quality nuclei with limited debris from frozen brain tissue.
Method
Nuclei were isolated from adult mouse brain tissue (RocklandTM, #MS-T004). The Frankenstein protocol was followed in order to obtain a nuclei suspension from the frozen brain tissue. Nuclei EZ lysis buffer from the Nuclei EZ Prep Nuclei Isolation kit (Sigma-Aldrich #NUC-101) was used. Nuclei were also filtered 2 times through a 30μm filter and remained on ice and at 4 degrees during the centrifugation steps. Keeping your sample cold is critical, failure to do so will result in degraded nuclei. Both the sample and sheath were kept chilled on the WOLF. The nuclei suspension was also stained with 7AAD to delineate the debris from nuclei. Sort gates were set on the WOLF to only include singlets and 7AAD+ nuclei. 200,000 nuclei were then sorted in a collection tube coated with 300μl of nuclei resuspension buffer (PBS + 1.0% BSA+0.2U/μl RNase Inhibitor). Nuclei purity was assessed by the WOLF. Nuclei quality was confirmed via microscopy after gentle centrifugation (300g at 5min) and staining with trypan blue.
Results
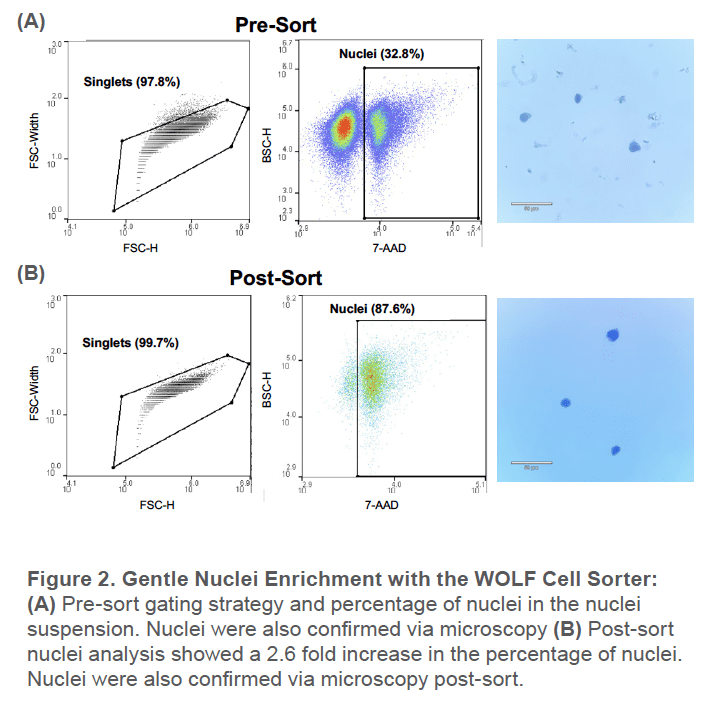
Pre-sort, around 33% of the nuclei suspension was nuclei while the remaining was unwanted cellular tissue. This was also reflected by the significant amount of debris observed under the microscope (Figure 2A). After sorting for the 7AAD+ singlet nuclei, the nuclei population increased to around 88%. There was also no debris observed under the microscope post-sort and the nuclei remain intact after gentle centrifugation (Figure 2B).
Conclusion
The WOLF Cell Sorter is a gentle microfluidic cell sorter that can gently and accurately sort nuclei while removing tissue and debris. In addition, gentle centrifugation after sorting still maintained high quality nuclei which demonstrates that this workflow is compatible for downstream genomic applications such as the 10x Genomics assays which require a concentrated input sample. In summary, this experiment demonstrates a robust workflow that optimizes nuclei purity without compromising nuclei quality.
For more information, visit nanocellect.com or email [email protected]
References
1. Luciano G Martelotto, Luciano Martelotto 2020. ‘Frankenstein’ protocol for nuclei isolation from fresh and frozen tissue for snRNAseq. protocols.io https://dx.doi.org/10.17504/protocols.io.bqxymxpw
2. Hu P, Fabyanic E, Kwon DY, Tang S, Zhou Z, Wu H. Dissecting Cell-Type Composition and Activity-Dependent Transcriptional State in Mammalian Brains by Massively Parallel Single-Nucleus RNA-Seq. Mol Cell. 2017 Dec 7;68(5):1006-1015.e7. doi: 10.1016/j.molcel.2017.11.017. PMID: 29220646; PMCID: PMC5743496.
3. Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, Regev A. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016 Aug 26;353(6302):925-8. doi: 10.1126/science.aad7038. Epub 2016 Jul 28. PMID: 27471252; PMCID: PMC5480621.
4. Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, Weitz DA, Rozenblatt-Rosen O, Zhang F, Regev A. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017 Oct;14(10):955-958. doi: 10.1038/nmeth.4407. Epub 2017 Aug 28. PMID: 28846088; PMCID: PMC5623139.
5. Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, Vijayaraghavan R, Wong J, Chen A, Sheng X, Kaper F, Shen R, Ronaghi M, Fan JB, Wang W, Chun J, Zhang K. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016 Jun 24;352(6293):1586-90. doi: 10.1126/science.aaf1204. PMID: 27339989; PMCID: PMC5038589.
6. Lacar B, Linker SB, Jaeger BN, Krishnaswami SR, Barron JJ, Kelder MJE, Parylak SL, Paquola ACM, Venepally P, Novotny M, O’Connor C, Fitzpatrick C, Erwin JA, Hsu JY, Husband D, McConnell MJ, Lasken R, Gage FH. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016 Apr 19;7:11022. doi: 10.1038/ncomms11022. Erratum in: Nat Commun. 2016 Jun 14;7:12020. Erratum in: Nat Commun. 2017 Mar 17;8:15047. PMID: 27090946; PMCID: PMC4838832.
APN – 025
