Increased Viability and Genomic Integrity of CRISPR-modified hiPS cells selected with WOLF Cell Sorter Microfluidic Technology
Background
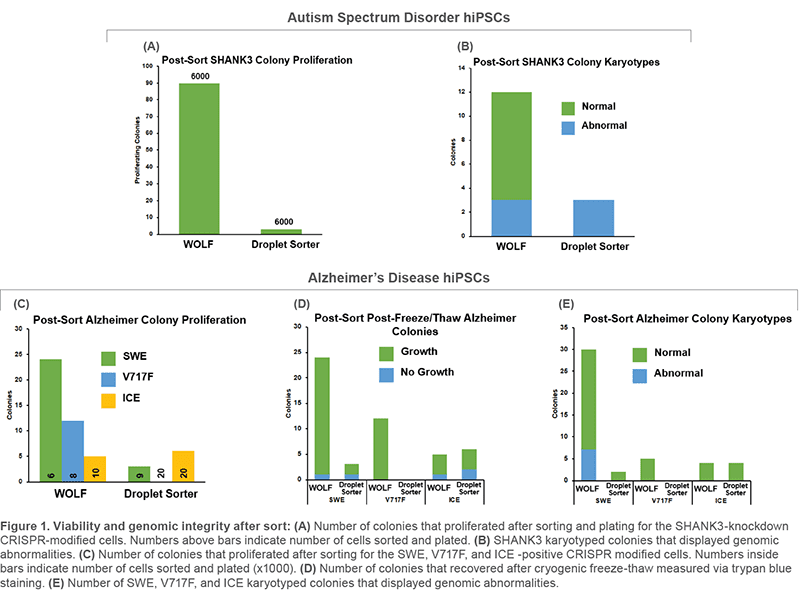
Human iPS cells (hiPSC) can be modified utilizing CRISPR/Cas9 gene editing technology to generate in vitro models of human diseases. Critical shortcomings in the generation of CRISPR-modified clonal lines are: 1) low viability/proliferation, and 2) genomic abnormalities following selection via fluorescent activated cell sorting. To address these issues, we compared microfluidic cell sorting (WOLF Cell Sorter, NanoCellect) to a traditional electrostatic droplet based cell sorter in Alzheimer’s Disease (AD) and Autism Spectrum Disorder (ASD) model cell lines. We evaluated colony formation and karyotypes after sorting.
Methods
Human fibroblasts were reprogrammed into hiPSC using retroviruses containing the four Yamanaka Factors. Resulting hiPSCs were then CRISPR-modified to knock-down AD- or ASD-related genes. GFP was used as reporter for selection. Cells were sorted in parallel with the WOLF and a traditional droplet-based cell sorter. After sorting, cells were plated on MEFs and grown for 1-2 weeks, picked, expanded, and frozen. Colonies that recovered after cryogenic storage were submitted for microarray karyotype analysis.
Results
ASD-related SHANK3 and AD-related SWE, V717F, and ICE cell lines displayed improvements in viability and genomic integrity following microfluidic sorting on the WOLF, compared to a traditional droplet cell sorter. The number of proliferating colonies, normal karyotypes, as well as overall freeze-thaw viability were all increased on the WOLF microfluidic platform compared to a droplet sorter.
APN-001