Gentle Sorting Accelerates Generation of Genome-Edited iPSCs
Introduction
CRISPR-edited iPSCs for in vitro disease models
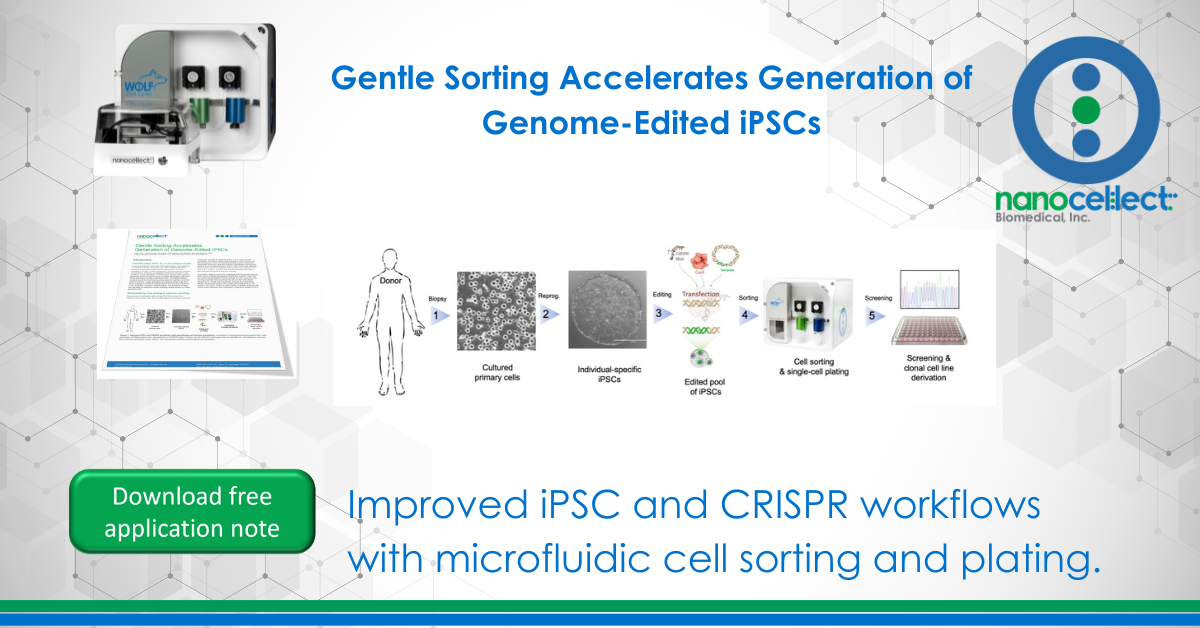
Induced pluripotent stem cell (iPSC) generation, from patients or healthy controls, combined with rapid CRISPR genome editing has greatly accelerated the generation of in vitro “disease-in-a-dish” model systems for studying human disease mechanisms. However, substantial limitations still exist in the workflow. In the derivation of iPSCs from patient cells, specific selection criteria could be used to isolate pluripotent cells from those that have not been reprogrammed. The differentiation process could be improved by using specific markers to enrich cells of interest. Lastly, selecting and plating single cells that have undergone CRISPR editing would improve the efficiency of the workflow. At each of these steps, low-pressure sorting can be used for selection while avoiding undue stress on these sensitive cells.
Reconsidering flow sorting to optimize workflow
Fluorescence activated cell sorting (FACS) is a common method of purifying cells from a heterogenous mixture, however it has been avoided for delicate iPSCs due to harsh sorting conditions. The shear stress, high pressures (up to 70 psi), and decompression shock of traditional droplet sorters have been associated with poor post-sort viability. Microfluidic mechanical sorters, such as NanoCellect’s WOLF® Cell Sorter have reduced shear stress due to microfluidic laminar flow, low pressure (<2 psi), and a droplet / aerosol-free mechanism that eliminates decompression shock to improve viability.
The WOLF detects and sorts unlabeled or fluorescently labeled cells within a sterile disposable microfluidic system. Cells can be sorted into standard microcentrifuge or 5 mL tubes for bulk sorting or sorted one-at-a-time into 96- or 384-well plates using the N1 Single Cell Dispenser module. By integrating sterile, gentle cell sorting and single cell dispensing in one system, the WOLF offers a viable option to greatly improve the monoclonal selection and outgrowth of genome-edited iPSCs.
Results
Monoclonal sorting and plating accelerate colony selection
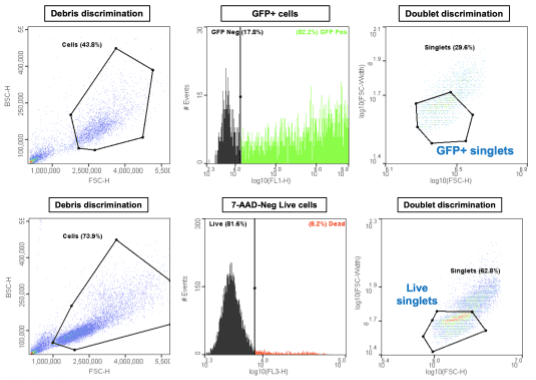
The WOLF yields a single cell plating efficiency of up to 93% which is significantly more efficient than non-sorting methods, such as limiting dilution. To evaluate the WOLF’s iPSC plating performance, a genome-edited iPSC line was cultured with mTeSR1 (STEMCELL Technologies) and StemFlex (Thermo Fisher Scientific) on hESC-qualified Matrigel plates (Corning). Cell-cluster passaging was performed using ReLeSR (STEMCELL Technologies) and single-cell dissociation was performed using Accutase. Unlabeled cells were sorted based on forward- and back-scatter properties to isolate singlets (Figure 2A), which were then dispensed into Matrigel-coated 96-well plates. Single iPSC clones were grown in StemFlex and were supplemented with 10μM ROCK inhibitor for the first 24 hours.
The WOLF shows improved monoclonal outgrowth
The microfluidic WOLF Cell Sorter and the conventional BD FACSAria II were each used to generate a 96-well plate of monoclonal cells to evaluate plating efficiency and outgrowth. The single-cell clones were allowed to grow and imaged to quantify colony growth. Though single cells were accurately dispensed into both, the cells in the Aria plate were no longer viable one week after plating. In contrast, the WOLF yielded outgrowth of 14 single cell colonies at the same timepoint (Figure 2B).
Sorting allows for selection of only viable, transfected cells
In addition to selecting and dispensing cells based on scatter properties, the WOLF Cell Sorter allows you to sort and dispense based on fluorescence detection, such as GFP expression and fluorescent viability stains. Fluorescent labels can be used with conventional plasmid transfections (Figure 3, top row), as well as with those utilizing the much more efficient CRISPR-Cas9 system. In addition, dead cells can be excluded with dyes, such as 7-AAD, that label cells with compromised cell membranes (Figure 3, bottom row). To minimize doublets, which is critical for single cell cloning, the software can be used to gate and sort only singlets.
Conclusion
Significant advances have demonstrated the scientific value of in vitro patient-cell-derived disease models. The increased use of these models requires improvements that reduce labor and increase efficiency in selection and cloning. Utilizing traditional FACS instruments to sort and isolate edited single cells has proven to yield very few or a complete lack of viable cells for outgrowth, rendering it an unusable option for delicate cell types. However, the WOLF Cell Sorter uses microfluidic sorting to overcome these challenges, resulting in reliable cloning for iPSCs. This high viability, disposable microfluidic technology reestablishes cell sorting as a viable option to advance the generation of engineering sensitive model cell lines.
APN-005


