Single-Cell Cloning for Cell Line Development
Introduction
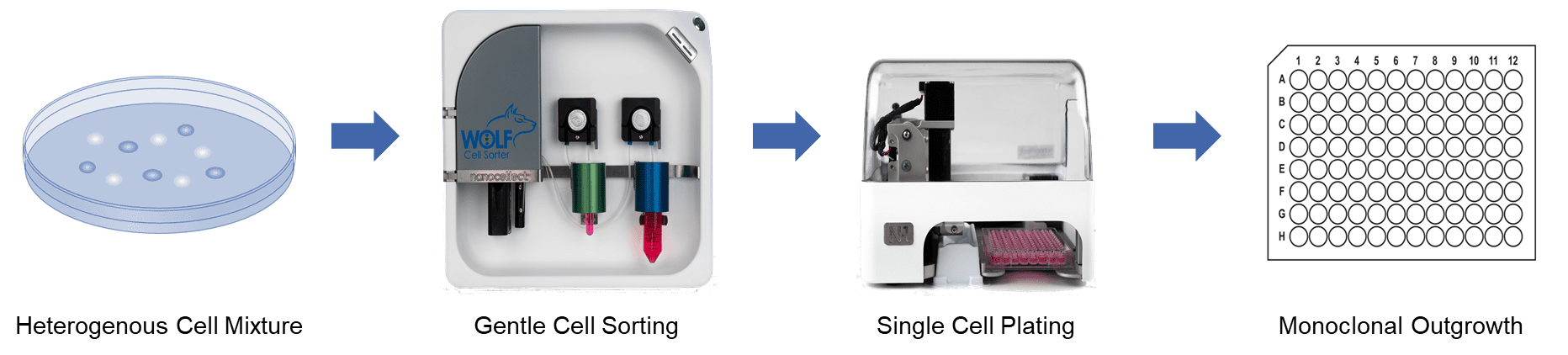
Generation of stable monoclonal cell lines is required for a wide variety of applications, including the development of biologics, such as therapeutic antibodies, or to create CRISPR-engineered cells for disease-in-a-dish models. While the demand for engineered cell lines has increased, production of monoclonal lines is still limited by the current technology for single-cell sorting and dispensing. Here we contrast the limitations of traditional technologies with newer technologies, such as microfluidic sorters and dispensers (WOLF® Cell Sorter, NanoCellect®, Inc.) that can analyze, sort and dispense single cells into 96- or 384-well plates with high viability and outgrowth. Here we highlight that both robust immortalized cell lines (e.g. CHO) and more sensitive cells can be used to generate monoclonal cell lines in a sterile format with little cellular stress.
Technologies for single-cell cloning need a boost
Traditional methods like laser capture microdissection (LCM) and colony picking by micromanipulator enable isolation of single cells, however their workflows are not compatible with living cells and/or are very low throughput. Using a classical technique, like limiting dilution, has been a go-to solution with minimal upfront resource requirements and gentle handling of cells. However, limiting dilution cannot positively select protein-producing cells or avoid dead cells, and is very inefficient, resulting in redundant downstream imaging and titer work to isolate monoclonal lines.
Fluorescence Activated Cell Sorting (FACS) methods allow analysis and sorting of specific cells with higher throughput than LCM, micromanipulation, or limiting dilution, with the ability to analyze multiple cell parameters (fluorescence and scatter) and directly sort into 96- or 384-well plates in minutes. Yet the traditional droplet-based cell sorting mechanism and fluidics system exposes cells to substantial shear stress and large, rapid changes in pressure (decompression shock) that can kill or injure cells.
This reduces outgrowth of cell colonies—especially in delicate, engineered lines or stem cells. Sample-to-sample contamination is also a major concern with traditional FACS because fluidic lines are not disposable and must be cleaned chemically. To address stress and sterility, cell printer technologies can plate single cells into plates, but these lack the analytical advantages of flow cytometry that allow the selection of living, single-cells that express a protein of interest to further increase plating efficiency.
The WOLF tackles traditional limitations
The WOLF Cell Sorter addresses challenges in sterility, viability and plating efficiency by combining microfluidic cell analysis and sorting with single-cell plating in the cell line development workflow. NanoCellect’s technology detects and sorts unlabeled or fluorescently labeled cells within a disposable microfluidic system, and then distributes the cells one-at-a-time to 96- or 384-well plates using the N1 Single Cell Dispenser module. A peristaltic pump gently pushes cells through microfluidic channels with low pressure (<2 PSI) and low shear stress. Robust laser-based excitation with sensitive PMT detection are used to identify cells that are gently sorted with a precise piezo actuator.
Microfluidic cartridges and tubing sets are provided in sterile, disposable kits to assure a clean environment for cell analysis, sorting and dispensing. Because of the microfluidic design, the WOLF uses only about 1% the amount of sheath fluid required by traditional cell sorters. This makes it feasible to use more expensive cell culture medium as sheath, if desired, to maintain cell health. Sorted cells are then dispensed via the N1 to a multi-well plate according to a user-defined layout. The tubing set connects magnetically to the N1 Single Cell Dispenser assuring end-to-end sterility of the fluid path. As each cell is analyzed and sorted within the WOLF’s cartridge, it immediately flows through to the N1 unit and is gently dispensed in 7 µL droplets that are “touched-off’ into each well. By integrating gentle cell sorting and single cell dispensing in a single system, cells have minimal handling and stress. This maximizes monoclonal viability, outgrowth and titer in cell line development, as demonstrated by the following data.
Results
High single-cell dispensing efficiency and outgrowth
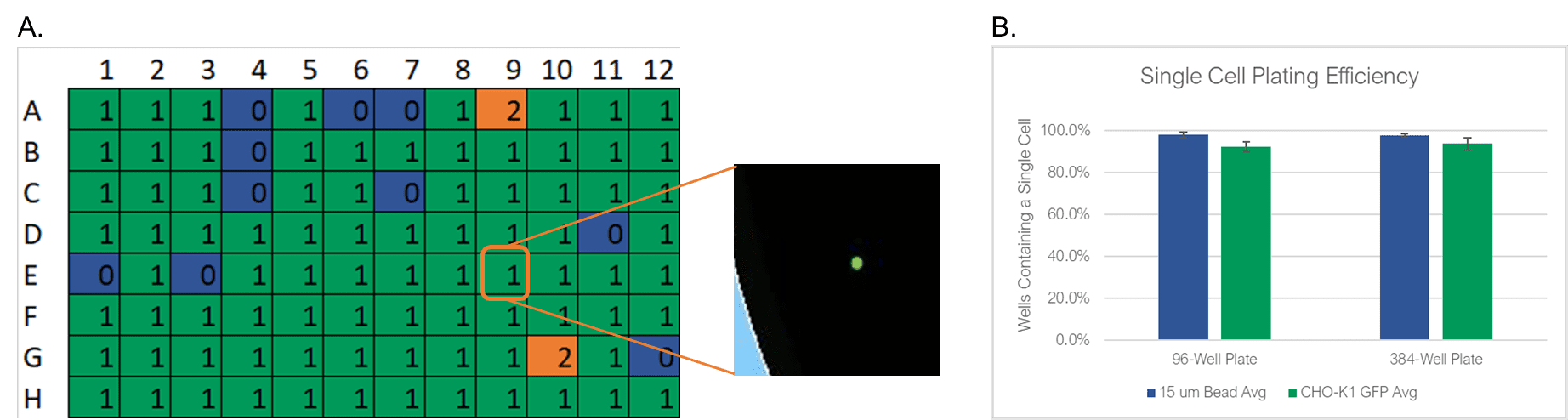
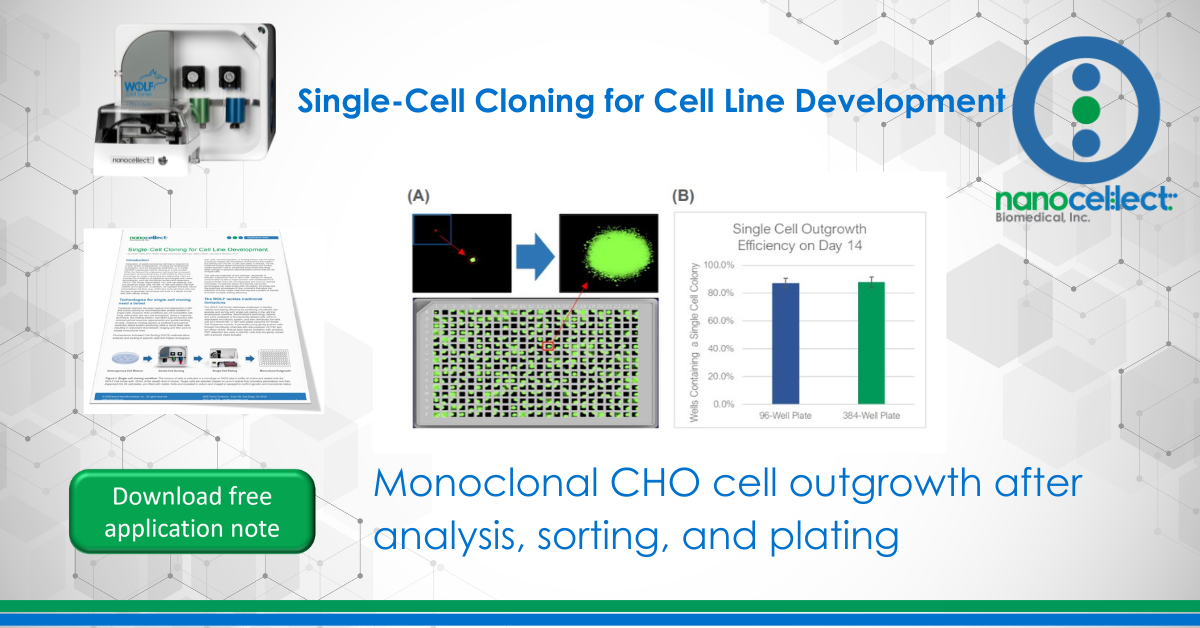
To test the WOLF sorting efficiency and accuracy, we sorted and dispensed 15 µm calibration beads (Dragon Green, Bangs Labs) and Chinese Hamster Ovary (CHO) cells, a cell line commonly used in the production of biologics. A stable GFP-expressing line of CHO cells was purchased (CHO-K1, ATCC), cultured and sorted on the WOLF by selecting high GFP expression and single-cells (doublet-discrimination). The WOLF sorted single, GFP+ cells into 96-well plates containing 200 µL/well culture media and into 384-well plates containing 50 µL/well culture media. Control plates were also tested with calibration beads. Fluorescent and brightfield images were taken and analyzed on Day 0 with a plate imager (NyOne, Synentech) with 4X magnification to measure singlet plating efficiency. Plates were then incubated at 37oC, 5% CO2 and imaged on day 14 to evaluate outgrowth. Three technical replicates were completed for 96-well (N=9) and 384-well (N=3) plates.
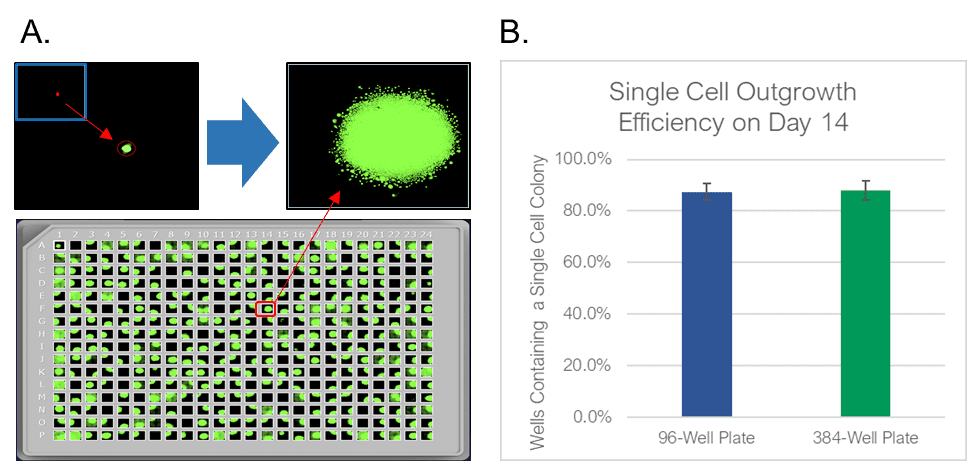
The WOLF Cell Sorter dispensed beads with a singlet fidelity of 98.1% on 96-well plates and 97.8% on 384-well plates. Similarly, dispensing CHO cells resulted in 92.5% and 93.8% single cells into 96- or 384-well plates, respectively (Figure 2). These results are comparable with traditional FACS technology, yet does so with lower mechanical stress. High shear stress commonly reduces colony outgrowth to below 30% with FACS. However, with the WOLF low-pressure microfluidics, >87% of wells had monoclonal outgrowth after 14 days in both 96- and 384-well plates (Figure 3).
Sensitive cells can be sorted to create >250% more colonies
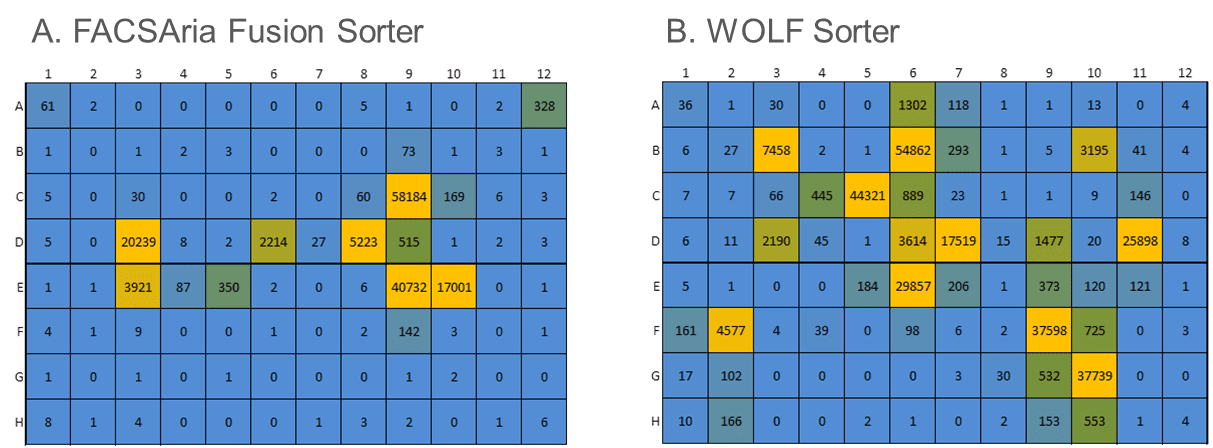
Cells engineered to produce large amounts of exogenous proteins can be very sensitive to manipulation and stress from traditional cell sorters. We compared the WOLF® to a traditional droplet-sorter in the lab of a Top-10 biopharma collaborator. Their existing workflow used a FACSAria Fusion (Becton Dickenson) to analyze, sort and plate a proprietary and highly-sensitive engineered Jurkat cell line with poor outgrowth. The WOLF Cell Sorter was evaluated in a side-by-side comparison to the FACSAria Fusion in analyzing, sorting and plating cells into 96-well plates, based on a specific fluorescent marker. Both plates were incubated for 2.5 weeks with one media addition midway through outgrowth. Cells from each of the wells were counted via flow cytometry based on a viability marker to evaluate colony growth efficiency.
Compared to the FACSAria Fusion, the WOLF sorter plated 20% more single cells (Figure 4). However, more importantly, after incubating 2.5 weeks, the WOLF sorted plates contained 2.6-fold more colonies (100 cells or more). The gentle sorting mechanism of the WOLF Cell Sorter greatly improved the viability of these sensitive cells, compared to traditional FACS, resulting in better monoclonal outgrowth.
Generating monoclonal, high-titer colonies
Inefficient generation of cell lines results in increased plating and subsequent downstream screening, such as quantification of protein production. In collaboration with an industry-leading developer of antibody therapeutics, we evaluated the ability of the WOLF to rapidly identify very high-titer antibody cell lines. To do so, we challenged the WOLF by sorting our collaborator’s most difficult-to-grow proprietary cells stained with a FITC-conjugated line-specific antibody.
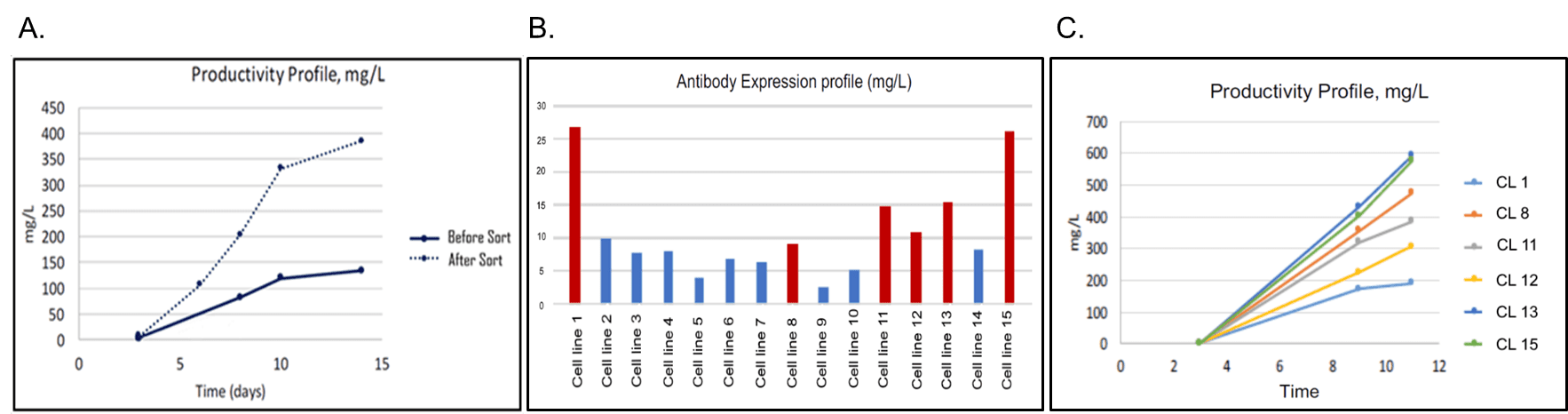
Engineered cells were bulk-sorted into a mini-pool of ~15,000 cells and evaluated for polyclonal antibody production in a 14-Day Fed-Batch Assay (Figure 5A). This simple bulk sort increased polyclonal productivity by more than 3-fold. Additionally, we attempted to improve monoclonal production. Sorted single cells were dispensed with the N1 into 96-well plates and incubated for 20 days with intermittent imaging to establish monoclonality. This yielded substantially more monoclonal lines than previously achieved by the collaborator. Once monoclonal lines reached 50% confluence, cells were transferred to a 24-well plate, put on a shaker, and, evaluated for antibody titer on Day 3 (Figure 5B). Six of the high-producing colonies were evaluated in an 11-Day Batch-Fed Assay for scaled-up antibody production. Two clones (1 and 15) demonstrated production up to 6-fold higher than unsorted cells (Figure 5C). These improvements in cell outgrowth and protein production save time and resources and are being applied by the collaborator to additional lines to improve titers and turnaround times.
Conclusion
Current technologies struggle to combine analysis, sorting and single-cell plating in an efficient, streamlined workflow for cell line development. Methods such as manual picking can effectively isolate cells of interest but are time consuming and low throughput. While technically easy and gentle, methods such as limiting dilution lack selection and therefore suffer from low efficiency. The WOLF combines multiparameter selection with gentle single-cell plating into a workflow that, with just 15 minutes of start-up, can efficiently generate monoclonal lines with high outgrowth and titers. The WOLF dispenses 96-well plates in 8 minutes with 92.5% efficiency and 87% outgrowth. Viability is highlighted by the ability of the WOLF to produce >2.5-fold more colonies when compared to traditional FACS technology. In summary, the microfluidic WOLF can improve cell line development workflows by improving sterility, viability, outgrowth, and titers.
APN-003