Attaining a sample quality that is sufficient for a sequencing experiment can be almost impossible when working with fragile cell or tissue samples. Furthermore, tissue dissociation methods can alter the transcriptome and cause significant cell death. Using nuclei instead of intact cells has grown
in popularity for downstream next-generation sequencing applications such as single nucleus RNA-Seq (snRNA-seq) and single-cell DNA sequencing. Using nuclei expands the use of samples and cell types that are difficult to isolate as single cells, such as neurons. However, nuclei isolation methods often contain a significant amount of debris in the suspension that cause clumping and poor sequencing results. Therefore, debris removal is a critical sample preparation step in generating robust sequencing results. One common way to reduce sample debris is by using a cell sorter; however, traditional high-pressure, droplet-based cell sorters can damage nuclei. The WOLF Cell Sorter is a gentle, microfluidic cell sorter that is capable of efficiently removing unwanted particles from samples with very low shear stress. Furthermore, the WOLF reduces biohazards with aerosol-free, disposable sorting cartridges that eliminate cross contamination between samples.
In this application note, we partnered with Invent Biotechnologies Inc. and Biostatus Ltd. to develop a quick and gentle method for isolating and sorting mammalian nuclei.
Invent Biotechnologies is known for developing novel protein extraction and cell fractionation kits. The Minute™ Nuclei isolation kit used in this study employs a proprietary filter cartridge-based technology and anti-aggregate buffer system for rapid isolation of single nuclei from fresh or frozen tissues. These kits are simple and easy to use and requires only a benchtop centrifuge. The whole protocol can be completed in less than 30 minutes, which is significantly faster than traditional nuclei isolation methods.
DRAQ7™ is a far-red fluorescent dye with high specificity for dsDNA. DRAQ7™ is cell impermeant, designed primarily as a reporter of cell death / late apoptosis. However, it rapidly labels isolated nuclei in a stoichiometric manner, according to DNA content, for sorting onwards to downstream molecular analysis e.g. snRNA-seq and snATAC-seq, as reported1. For added convenience, DRAQ7 DROP & GO™, employed in this work, is a dropper bottle presentation of DRAQ7™, used directly from the bench.
Human embryonic kidney 293 (HEK293) cells and mouse brain tissue (25mg) (Rockland Immunochemicals, Inc.) were processed to a nuclei suspension using the Invent Biotechnologies Minute™ Detergent-Free Nuclei Isolation Kit (#NI-024) and Minute™ Single Nucleus Isolation Kit
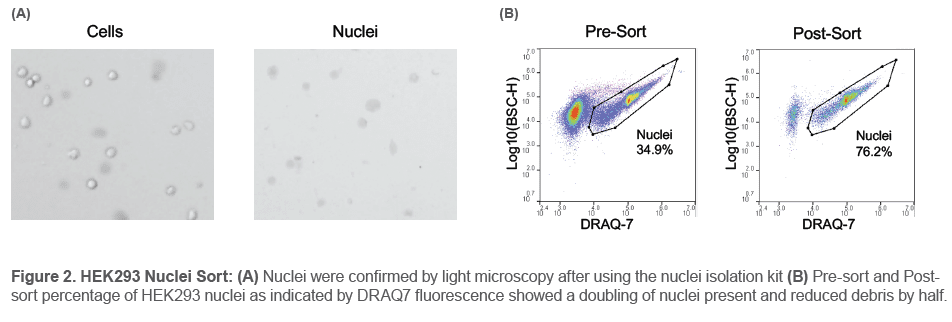
for Neuronal Tissues/Cells (#BN-020) respectively. Nuclei were then confirmed using Trypan blue on the Countess™ II Automated Cell Counter (Figure 2A). The nuclei cell suspension was then diluted to 200-300 events/μL with PBS + 0.1% BSA and filtered through a 37 μm filter. Nuclei were then stained with DRAQ7 DROP & GO™ at 2 drops per 0.5 mL and incubated on ice for 10 mins. An unstained nuclei sample was used for a negative control. Nuclei suspensions were then analyzed and
sorted on the WOLF. Sort gates were set to exclude debris by gating out DRAQ7™ negative events (Figures 2-3).
Nuclei could be identified by looking at the BSC-H/DRAQ7 parameters. Discrete amounts of DNA content could also be identified based on the different intensities of DRAQ7™ fluorescence. From the HEK293 nuclei suspension, 35% of the population was identified as nuclei before sorting. After sorting for the DRAQ7+ nuclei population, the content of nuclei was more than doubled to over 76% (Figure 2B).
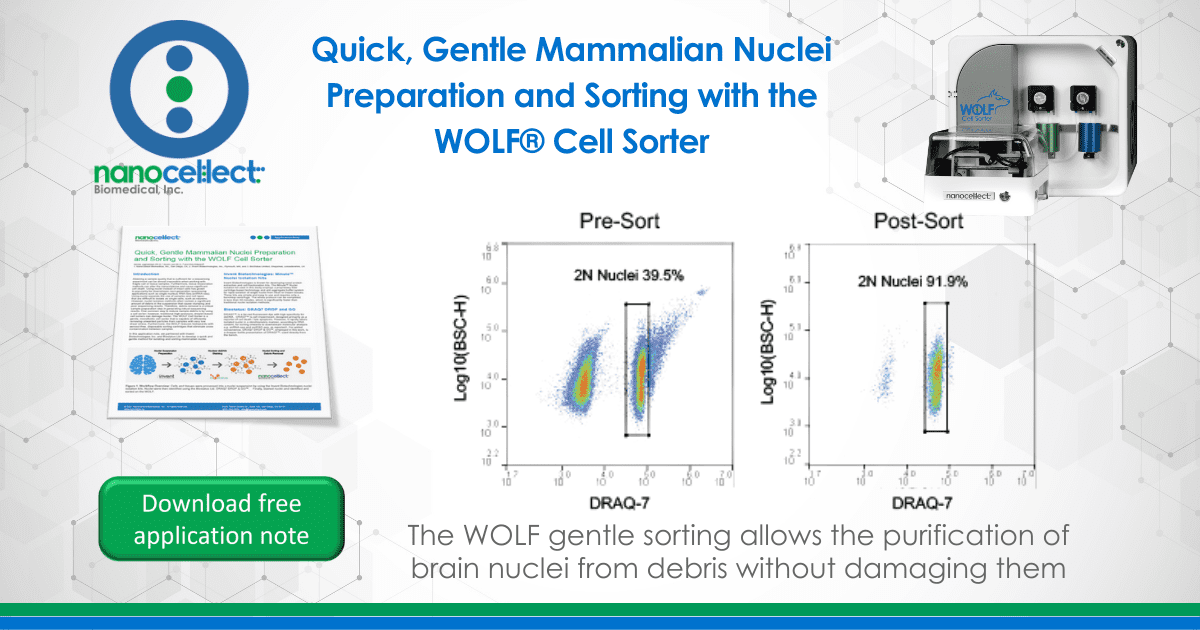
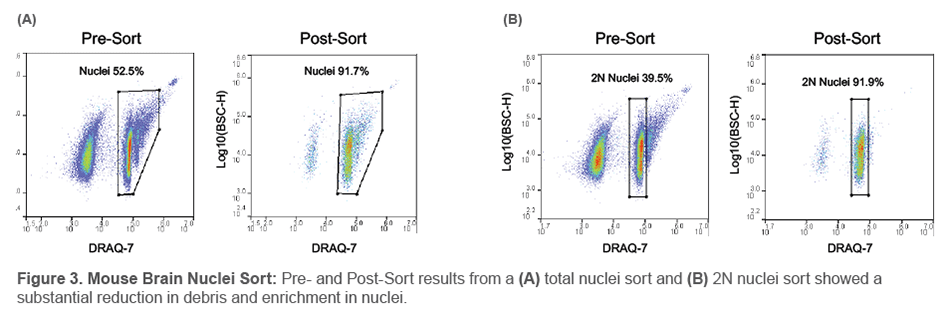
Next, mouse brain tissue was processed for nuclei suspension and stained with DRAQ7 DROP & GO™. Pre-Sort analysis showed that half of the sample was nuclei while the other half was contaminating debris. After using the WOLF to sort for only the DRAQ7+ nuclei population, we saw a significant reduction in the amount of debris and enriched the nuclei population to nearly 92%. Furthermore, some research studies may only be interested in a certain type of nuclei, so a 2N nuclei sort was also conducted on the WOLF. Results from this sort also showed an enrichment of 92% 2N nuclei.
Nuclei from both cells and tissue were successfully isolated and sorted by using combined novel technologies. By using the Invent Biotechnologies nuclei isolation kit, sample preparation only took 30 minutes and the nuclei remained intact. Moreover, no clogs were observed during the sort on
the WOLF. Furthermore, successful staining was observed with Biostatus’ DRAQ7 DROP & GO™ in an incubation time of only 10 minutes. In conclusion, results from these experiments demonstrate a gentle and quick workflow to isolate and sort nuclei from cells and tissue for downstream applications such as snRNA-Seq.
APN-023





