Single-cell RNA-sequencing has led to many discoveries, such as the detection of rare cell populations, microbial
diversity, and cancer mutations. The WOLF Cell Sorter and N1 Single Cell Dispenser, developed by NanoCellect, is a novel microfluidic-based cell sorter compatible with several RNASequencing platforms. At less than 2 psi, the WOLF is more gentle than conventional cell sorters, enabling healthier cells and higher RNA integrity post-sort. Low shear stress during cell sorting avoids potential gene expression changes induced by traditional sorters. In addition, the WOLF excels at excluding dead cells and debris; therefore maximizing the data generated per dollar spent on sequencing reagents and analysis time. Furthermore, the WOLF’s microfluidic cartridges are completely
disposable, everything the sample and sheath fluid touches is sterile and free from sample to sample contamination enabling more accurate sequencing results. In addition, with 5 parameters of detection, the WOLF provides higher rates of singlet detection and live/dead discrimination compared to cell printers and limiting dilution.
When isolating a target population for the Chromium Controller workflow, it is critical to maximize cell viability. It is recommended that cell populations contain more than 90% viable cells prior to loading your sample into the Chromium Controller. Using the WOLF upstream of loading your sample into the Chromium Controller allows isolation of the target cell population while removing dead cells, debris and doublets. Here, we demonstrate the genomic sequencing benefits of using the WOLF prior to loading your sample into the Chromium Controller.
Comparing Unsorted and WOLF Sorted PBMCs in the 10x Genomics Workflow
In order to determine the genomic benefits of sorting cells through the WOLF prior to 10x library construction, PBMCs were isolated from human blood using a density gradient, and then split into two samples: 1) an unsorted sample and 2) a sample sorted on the WOLF. In the unsorted sample, 10,000 cells were prepared to a concentration of 1000 cells/μL and loaded into the Chromium Controller. In the WOLF sorted sample, we stained cells with Propidium Iodine (PI) to identify dead cells and then performed viable cell sorting on the WOLF.
Sorting gates were then set to exclude PI+ dead cells, debris and doublets. 10,000 of these sorted cells were prepared at 1000 cells/μL and loaded into the Chromium Controller (Figure 1). Both samples were resuspended in PBS + 2% FBS. RNA libraries were then generated using the Chromium Single Cell 3’ v3 Kit and sequenced on the Illumina HiSeq 4000. Sequencing analysis done using the Cell Ranger pipeline in collaboration with the UCSD Genomics Core.
Using the WOLF Cell Sorter increases library complexity and quality
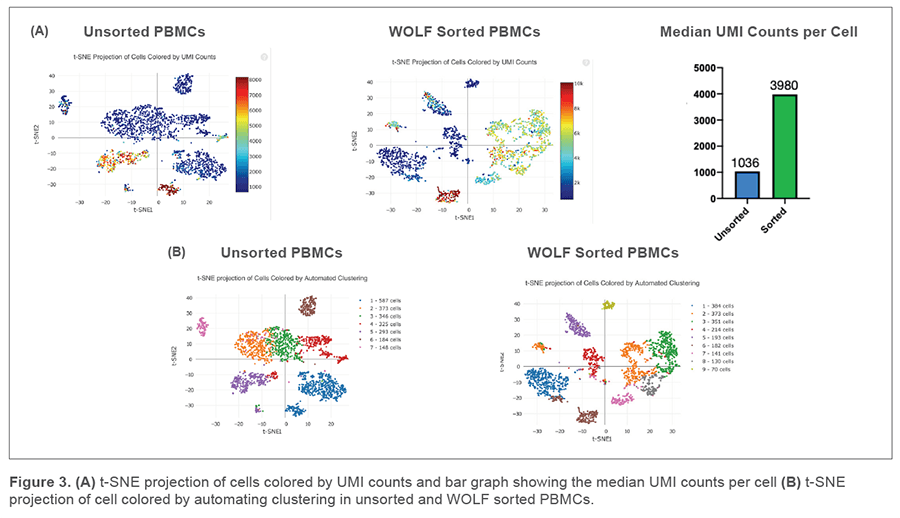
The Cell Ranger performance sequencing summary showed 2,256 cells captured in the unsorted sample and 2,038 cells captured in our WOLF sorted sample. This was consistent with our expected recovery due to routine handling and the Chromium Controller capture rate of up to 65%. We detected over twice the median genes per cell in our WOLF sorted sample (1,101 genes) compared to our unsorted sample (456 genes) (Figure 2A). In addition, there were more total genes in the WOLF sorted sample (Figure 2B). Having a higher median of genes per cell and more total genes detected both indicate higher library complexity in our WOLF sorted sample. We also examined the fraction of reads that were not associated with a cell. There was nearly twice the amount of cell-free RNA contamination in the unsorted sample, relative to the WOLF sorted sample (Figure 2C). These results highlight that there was decreased contamination from dead cells in the WOLF sorted sample.
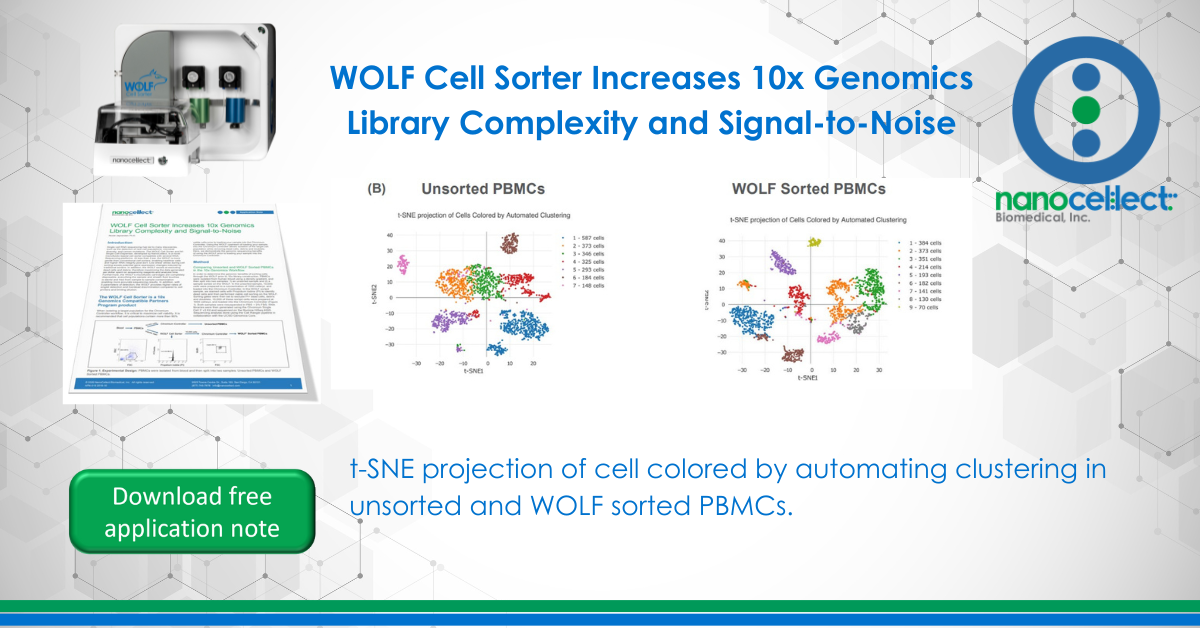
More UMIs per cell and distinct cell populations were identified in the WOLF sorted sample
T-distributed Stochastic Neighbor Embedding (t-SNE) plots were generated on our unsorted and WOLF sorted sample through the Cell Ranger pipeline. t-SNE projection of cells colored by UMI counts shows that there were substantially more cells with low UMI counts per cell (blue dots) in the unsorted sample compared to the WOLF sorted sample. Specifically, there were approximately 3 times more UMIs detected per cell in our WOLF sorted sample. This reflects a higher number of RNA transcripts per cell detected in the WOLF sorted sample (Figure 3A, next page). t-SNE projection of cells colored by automated clustering produced only 7 clusters of cells in the unsorted sample, relative to 9 clusters of cells in the WOLF sorted sample. Furthermore, clusters were more distinct in the WOLF sorted sample (Figure 3B). These results further indicate that background contamination was reduced and a higher sequencing depth was achieved in the WOLF sorted sample.
The quality of sequencing results is directly dependent on the quality of the sample that is prepared. The WOLF Cell Sorter serves as a robust method to isolate target cells while removing dead cells, debris and doublets. In comparing 10x Genomics sequencing results from unsorted or WOLF sorted samples, the WOLF improved sample input for the Chromium Controller and improved sequencing results. Specifically, library complexity These results demonstrate that the WOLF is compatible with the 10x Genomics workflow and advantageous for obtaining higher quality results. In summary, the WOLF serves as a valuable tool to obtain optimal sequencing results while
allowing more data per dollar spent on genomic reagents.
APN-019





